solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1fmta1 ( 207 ) lskeeArId---WslsAaqLercIrafn-pwPMSwLeieg---qpVkVwk
d1xg9a4 ( 165 ) rpeqSeIs---pdfdlekiydyIrldgegyprAFiky-g---yrlefsr
d2blna1 ( 204 ) ddsfLe---WhkpAsvLhnmVravadpwpGAfSyvgn---qkFTVws
d2bw0a1 ( 204 ) qkketAkIn---WdqpAeaIhnwIrgNd-kvpGAWTeAce---qkLTFfn
d3rfoa2 ( 205 ) ikreqekId---WtktGeeVynhIrgln-pwpVAyTtLag---qvVkVww
d3ubya1 ( 84 ) trlglefFdqpAvpLAraFLGq-----vLVRrlpngteLrGrIVE
aaaaaaaa bbb bbbbbbb
60 70 80 90 100
d1fmta1 ( 250 ) Asvid---tatnaa-----------------pGtIleAnk----qGIqVA
d1xg9a4 ( 210 ) Askn----------------------------------------------
d2blna1 ( 245 ) Srvhp---haskaq-----------------pGSVisva------pLlIA
d2bw0a1 ( 247 ) StlntsglvpegdaLpIp------------gAhrpGvVtk----aGLILf
d3rfoa2 ( 248 ) Gekvp---vtksae-----------------aGtIvaiee----dGFVVA
d3ubya1 ( 124 ) TEAylGpe--DeaAhSrggrqtprNrGMfmkpGTLYVyiiygmyfcMNIS
bb bb bbb
110 120 130 140 150
d1fmta1 ( 276 ) Tgd--giLnLlsLqPag--------------kkamsAqdLlnsrr-ewFv
d1xg9a4 ( 215 ) -----gkiiadveiieg
d2blna1 ( 269 ) Cgd--GALeIvtGqagd--------------gitmqGsqLAqtLg---Lv
d2bw0a1 ( 281 ) Gndd-kmLLVknIqled--------------gkmilAsnFfk
d3rfoa2 ( 274 ) Tgne-tGVkIteLqpsg--------------kkr-sCsqflrgtk---pe
d3ubya1 ( 172 ) SqgdGACVlLRALePlegletMrqlrstkdreLCsgpskLCqALaInksF
bbbbbbbbb
160 170 180 190 200
d1fmta1 ( 309 ) pgnrLv
d1xg9a4
d2blna1 ( 300 ) qgsrL
d2bw0a1
d3rfoa2 ( 306 ) igtkLg
d3ubya1 ( 232 ) DqrdLaqdeaVwLerGplpsepaVVaAaRvgvagewarkplRFYvrgspw
d1fmta1
d1xg9a4
d2blna1
d2bw0a1
d3rfoa2
d3ubya1 ( 285 ) VSvvdrvae
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| Methionyl-tRNAfmet formyltransferase, C-terminal domain | Post formyltransferase domain | Escherichia coli [TaxId: 562] | 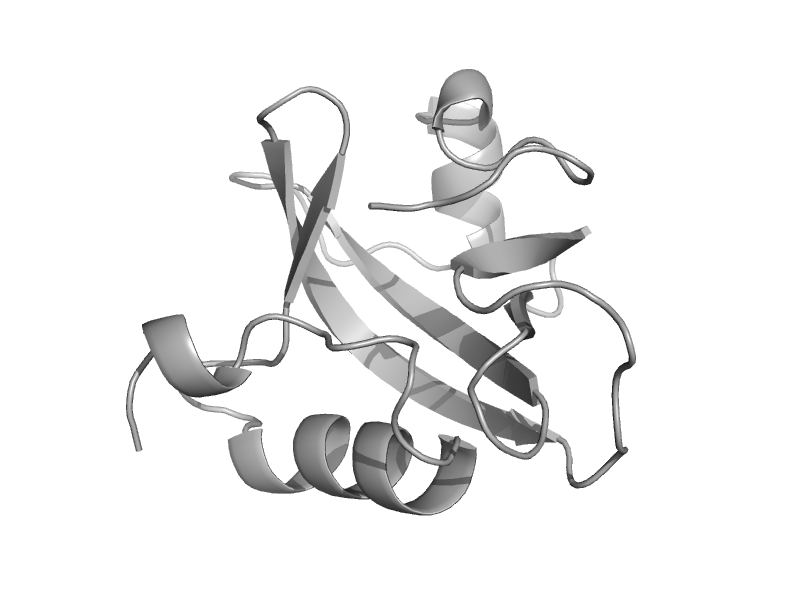 | view | |
| Methionyl-tRNAfmet formyltransferase, C-terminal domain | Post formyltransferase domain | Clostridium thermocellum [TaxId: 1515] | 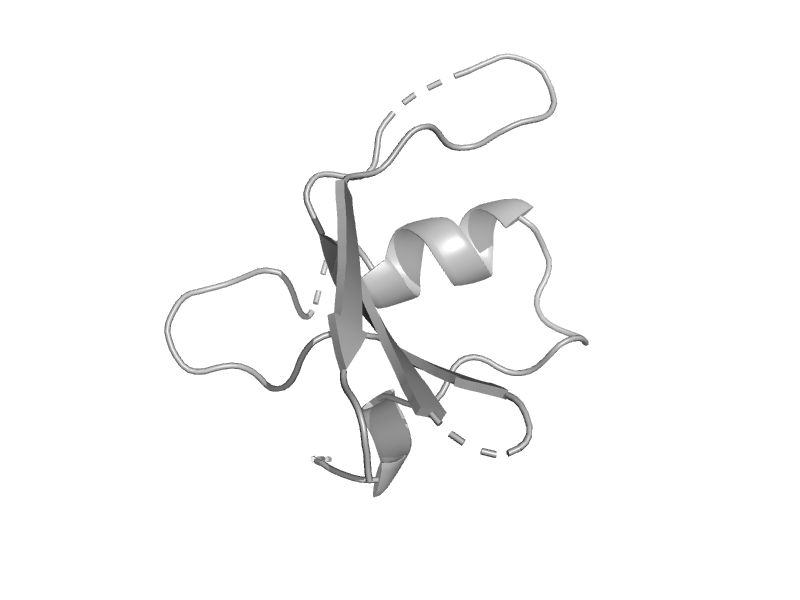 | view | |
| Polymyxin resistance protein ArnA, domain 2 | Post formyltransferase domain | Escherichia coli [TaxId: 562] | 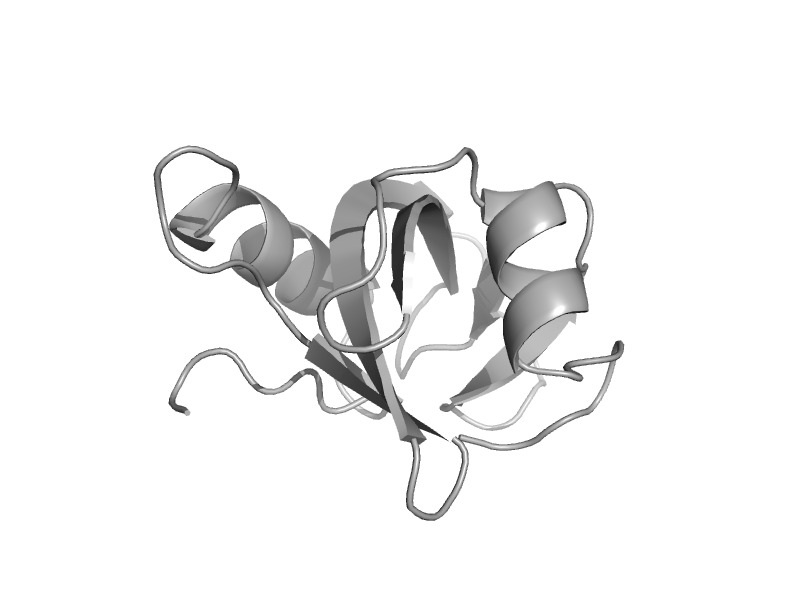 | view | |
| 10-formyltetrahydrofolate dehydrogenase domain 2 | Post formyltransferase domain | Human (Homo sapiens) [TaxId: 9606] | 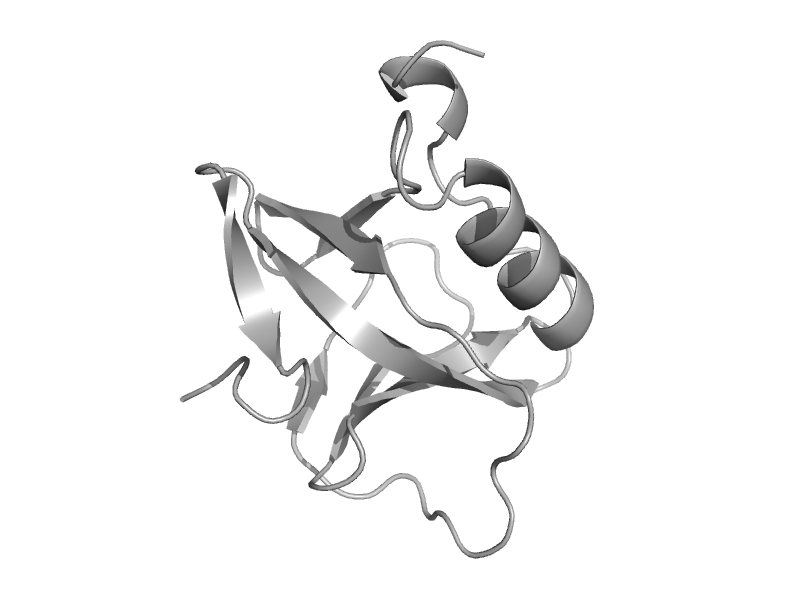 | view | |
| automated matches | automated matches | Anthrax bacillus (Bacillus anthracis) [TaxId: 1392] | 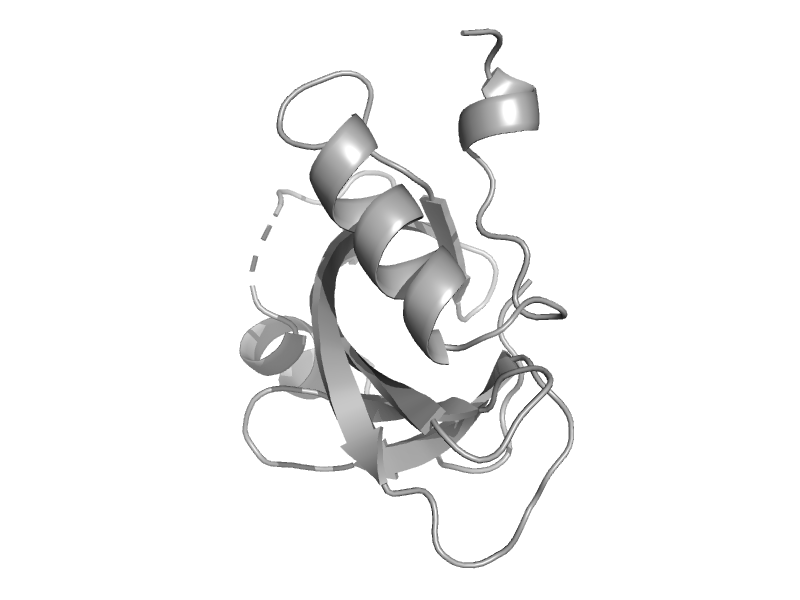 | view | |
| automated matches | 3-methyladenine DNA glycosylase (AAG, ANPG, MPG) | Human (Homo sapiens) [TaxId: 9606] | 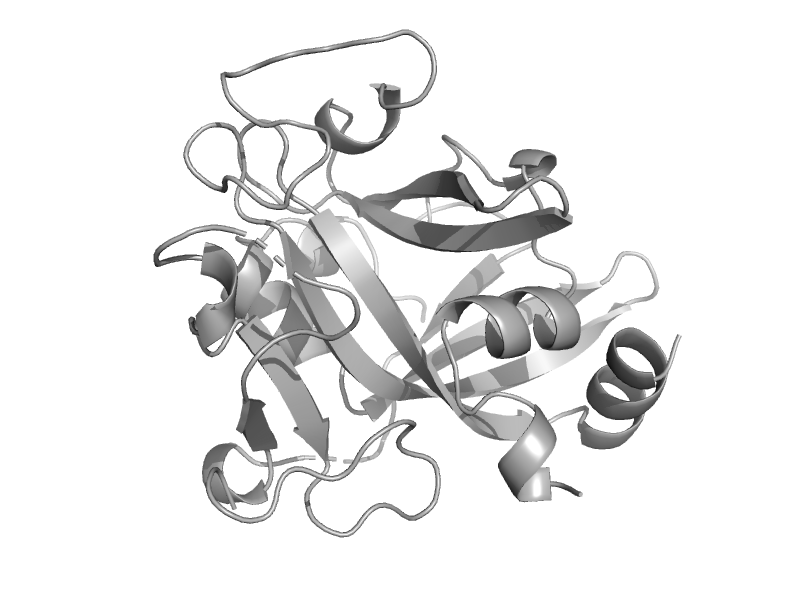 | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | 150 | 160 | 170 | 180 | 190 | 200 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L | S | K | E | E | A | R | I | D | - | - | - | W | S | L | S | A | A | Q | L | E | R | C | I | R | A | F | N | - | P | W | P | M | S | W | L | E | I | E | G | - | - | - | Q | P | V | K | V | W | K | A | S | V | I | D | - | - | - | T | A | T | N | A | A | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | P | G | T | I | L | E | A | N | K | - | - | - | - | Q | G | I | Q | V | A | T | G | D | - | - | G | I | L | N | L | L | S | L | Q | P | A | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | K | K | A | M | S | A | Q | D | L | L | N | S | R | R | - | E | W | F | V | P | G | N | R | L | V | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | R | P | E | Q | S | E | I | S | - | - | - | P | D | F | D | L | E | K | I | Y | D | Y | I | R | L | D | G | E | G | Y | P | R | A | F | I | K | Y | - | G | - | - | - | Y | R | L | E | F | S | R | A | S | K | N | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | K | I | I | A | D | V | E | I | I | E | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | D | D | S | F | L | E | - | - | - | W | H | K | P | A | S | V | L | H | N | M | V | R | A | V | A | D | P | W | P | G | A | F | S | Y | V | G | N | - | - | - | Q | K | F | T | V | W | S | S | R | V | H | P | - | - | - | H | A | S | K | A | Q | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | P | G | S | V | I | S | V | A | - | - | - | - | - | - | P | L | L | I | A | C | G | D | - | - | G | A | L | E | I | V | T | G | Q | A | G | D | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | I | T | M | Q | G | S | Q | L | A | Q | T | L | G | - | - | - | L | V | Q | G | S | R | L | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Q | K | K | E | T | A | K | I | N | - | - | - | W | D | Q | P | A | E | A | I | H | N | W | I | R | G | N | D | - | K | V | P | G | A | W | T | E | A | C | E | - | - | - | Q | K | L | T | F | F | N | S | T | L | N | T | S | G | L | V | P | E | G | D | A | L | P | I | P | - | - | - | - | - | - | - | - | - | - | - | - | G | A | H | R | P | G | V | V | T | K | - | - | - | - | A | G | L | I | L | F | G | N | D | D | - | K | M | L | L | V | K | N | I | Q | L | E | D | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | K | M | I | L | A | S | N | F | F | K | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| I | K | R | E | Q | E | K | I | D | - | - | - | W | T | K | T | G | E | E | V | Y | N | H | I | R | G | L | N | - | P | W | P | V | A | Y | T | T | L | A | G | - | - | - | Q | V | V | K | V | W | W | G | E | K | V | P | - | - | - | V | T | K | S | A | E | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | A | G | T | I | V | A | I | E | E | - | - | - | - | D | G | F | V | V | A | T | G | N | E | - | T | G | V | K | I | T | E | L | Q | P | S | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | K | K | R | - | S | C | S | Q | F | L | R | G | T | K | - | - | - | P | E | I | G | T | K | L | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | T | R | L | G | L | E | F | F | D | Q | P | A | V | P | L | A | R | A | F | L | G | Q | - | - | - | - | - | V | L | V | R | R | L | P | N | G | T | E | L | R | G | R | I | V | E | T | E | A | Y | L | G | P | E | - | - | D | E | A | A | H | S | R | G | G | R | Q | T | P | R | N | R | G | M | F | M | K | P | G | T | L | Y | V | Y | I | I | Y | G | M | Y | F | C | M | N | I | S | S | Q | G | D | G | A | C | V | L | L | R | A | L | E | P | L | E | G | L | E | T | M | R | Q | L | R | S | T | K | D | R | E | L | C | S | G | P | S | K | L | C | Q | A | L | A | I | N | K | S | F | D | Q | R | D | L | A | Q | D | E | A | V | W | L | E | R | G | P | L | P | S | E | P | A | V | V | A | A | A | R | V | G | V | A | G | E | W | A | R | K | P | L | R | F | Y | V | R | G | S | P | W | V | S | V | V | D | R | V | A | E |
| Member | Chain | UniProtKB | GO term |
|---|---|---|---|
| d1fmta1 | A | P23882 | GO:0003824 |
| d1fmta1 | A | P23882 | GO:0004479 |
| d1fmta1 | A | P23882 | GO:0005829 |
| d1fmta1 | A | P23882 | GO:0006412 |
| d1fmta1 | A | P23882 | GO:0006413 |
| d1fmta1 | A | P23882 | GO:0009058 |
| d1fmta1 | A | P23882 | GO:0016740 |
| d1fmta1 | A | P23882 | GO:0019988 |
| d1fmta1 | A | P23882 | GO:0071951 |
| d1fmta1 | A | P23882 | GO:0003824 |
| d1fmta1 | A | P23882 | GO:0004479 |
| d1fmta1 | A | P23882 | GO:0005829 |
| d1fmta1 | A | P23882 | GO:0006412 |
| d1fmta1 | A | P23882 | GO:0006413 |
| d1fmta1 | A | P23882 | GO:0009058 |
| d1fmta1 | A | P23882 | GO:0016740 |
| d1fmta1 | A | P23882 | GO:0019988 |
| d1fmta1 | A | P23882 | GO:0071951 |
| d2blna1 | A | P77398 | GO:0003824 |
| d2blna1 | A | P77398 | GO:0009058 |
| d2blna1 | A | P77398 | GO:0003824 |
| d2blna1 | A | P77398 | GO:0009058 |
| d2bw0a1 | A | O75891 | GO:0003824 |
| d2bw0a1 | A | O75891 | GO:0009058 |
| d2bw0a1 | A | O75891 | GO:0003824 |
| d2bw0a1 | A | O75891 | GO:0009058 |
