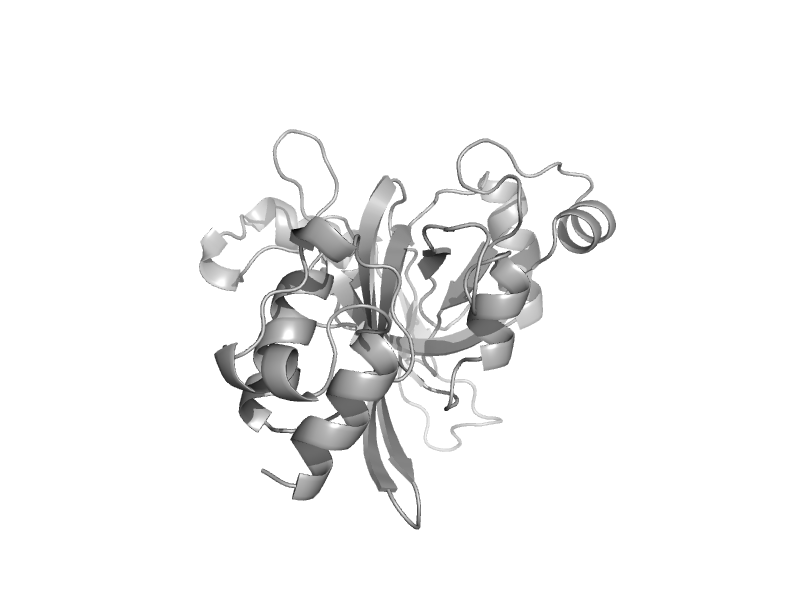
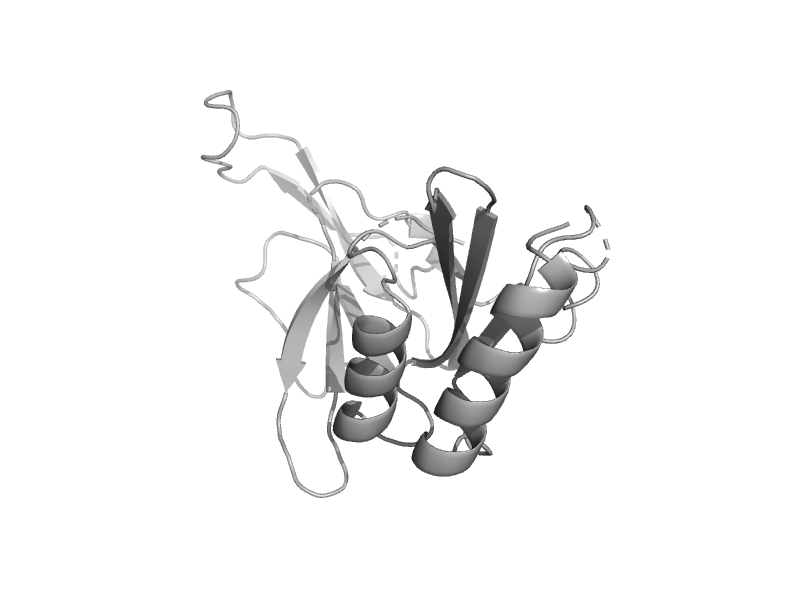
solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d3a9sa2 ( 361 ) aqIfAdVrtywspeaVkrvtgytLegraanGIIHLInsgaaaldGTGeQt
d4f2da2 ( 329 ) tSf-edytyhfek--------------gndLVlGShl-eVcpsIAve--
bbbbb bbbb 333
60 70 80 90 100
d3a9sa2 ( 411 ) kdgkpVIkpyyeltdeDikkCleaTqfrpasteyfr-gggystdFlTkGg
d4f2da2 ( 363 ) -----------------------kpildvqhlgiggkddparliFnTqt-
bbbb bbbb
110 120 130 140 150
d3a9sa2 ( 460 ) MpVTIsrlnivkglgpVlQIAeGyTvdLpeevhdvLdkrtdptwPTTWFv
d4f2da2 ( 389 ) gpAiVaSlidl-gdryrlLVNcIdTvktph--slp-----klpvANalWk
bbbbbbbbb bbbbbbbbbb bbbb
160 170 180 190 200
d3a9sa2 ( 510 ) PnltgegaFkdVysVmnnW----ganhCsIsyghiGadlItlASilriPV
d4f2da2 ( 431 ) Aqp-------dLptAseAwilAgGahhtvFShalnlnd-rqfAe-hdiEi
b aaaaaa bbb b
210 220 230
d3a9sa2 ( 556 ) nMhNVpeekIfrpdaWsmfgtkdlegADyrACkkl
d4f2da2 ( 474 ) tvIdn---dtr-lpafkd-----alrwnevyygf
bb aaaaaaaaa
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | 150 | 160 | 170 | 180 | 190 | 200 | 210 | 220 | 230 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | Q | I | F | A | D | V | R | T | Y | W | S | P | E | A | V | K | R | V | T | G | Y | T | L | E | G | R | A | A | N | G | I | I | H | L | I | N | S | G | A | A | A | L | D | G | T | G | E | Q | T | K | D | G | K | P | V | I | K | P | Y | Y | E | L | T | D | E | D | I | K | K | C | L | E | A | T | Q | F | R | P | A | S | T | E | Y | F | R | - | G | G | G | Y | S | T | D | F | L | T | K | G | G | M | P | V | T | I | S | R | L | N | I | V | K | G | L | G | P | V | L | Q | I | A | E | G | Y | T | V | D | L | P | E | E | V | H | D | V | L | D | K | R | T | D | P | T | W | P | T | T | W | F | V | P | N | L | T | G | E | G | A | F | K | D | V | Y | S | V | M | N | N | W | - | - | - | - | G | A | N | H | C | S | I | S | Y | G | H | I | G | A | D | L | I | T | L | A | S | I | L | R | I | P | V | N | M | H | N | V | P | E | E | K | I | F | R | P | D | A | W | S | M | F | G | T | K | D | L | E | G | A | D | Y | R | A | C | K | K | L |
| - | T | S | F | - | E | D | Y | T | Y | H | F | E | K | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | N | D | L | V | L | G | S | H | L | - | E | V | C | P | S | I | A | V | E | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | K | P | I | L | D | V | Q | H | L | G | I | G | G | K | D | D | P | A | R | L | I | F | N | T | Q | T | - | G | P | A | I | V | A | S | L | I | D | L | - | G | D | R | Y | R | L | L | V | N | C | I | D | T | V | K | T | P | H | - | - | S | L | P | - | - | - | - | - | K | L | P | V | A | N | A | L | W | K | A | Q | P | - | - | - | - | - | - | - | D | L | P | T | A | S | E | A | W | I | L | A | G | G | A | H | H | T | V | F | S | H | A | L | N | L | N | D | - | R | Q | F | A | E | - | H | D | I | E | I | T | V | I | D | N | - | - | - | D | T | R | - | L | P | A | F | K | D | - | - | - | - | - | A | L | R | W | N | E | V | Y | Y | G | F | - |
| Member | Chain | UniProtKB | GO term |
|---|