solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1cb8a2 ( 600 ) pkvlantnqlqAvYhqqldmVqAiFytagklsVagieIeTdkpCaVlIk
d1egua2 ( 815 ) s-sliennetlqSvydakqgvWgIvKyddsvstIsn-qFqVlkrgvYtIr
d1f1sa3 ( 912 ) evellensskqqViydknsqtwaViKhdnqeSlInn-qFkMnkaglYlVq
d1hn0a3 ( 900 ) yqvlrkdkdVhIildklsnvTgYaFyqpasIed-kwIkkVnkpAiVmTh
d1rwaa2 ( 645 ) kysvirndatAqSvefktakttaAtFwk--pgmAg--dLGAsgpAcVvfs
d1x1ia2 ( 660 ) paieivntsgvqSvkektlglvgAnFwtdttqtAd--LItSnkkAsVmTr
bbbbb bbbbbb bbbbbb bbb bbb bbbbbb
60 70 80 90 100
d1cb8a2 ( 649 ) hIn--gqvIwAaDplq--------kektAvLsir-dl-------------
d1egua2 ( 863 ) keg-deykIaYynpetqesapdqeVFkkq
d1f1sa3 ( 961 ) kvg-ndyqnvyyqpqtmtktdqlai
d1hn0a3 ( 948 ) rqk-dtLiVsAvTpdlnmtrqkaAtpvtInVtIn-gkwqsadknseVkyq
d1rwaa2 ( 691 ) rhg-neLsLaVsEptqkaa--------gLtLtLPegtWssvlega-gtlg
d1x1ia2 ( 709 ) eiaderLeAsVsDptqann-------gtIaIeLa-rsAegysadpgItVt
b bbbbbb
110 120 130
d1cb8a2 ( 676 ) kt-gktnrvkIdFpqqefagatvel
d1egua2
d1f1sa3
d1hn0a3 ( 996 ) vs-gdnTeLtFtSy----fgipqeIkLsplp
d1rwaa2 ( 731 ) tdadgrStLtLdTt--glsgktklIkLkr
d1x1ia2 ( 751 ) qla-ptIkFtVnVn--gAkgksfhAsFqlg
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| Chondroitinase AC | Hyaluronate lyase-like, C-terminal domain | Pedobacter heparinus (Flavobacterium heparinum) [TaxId: 984] | 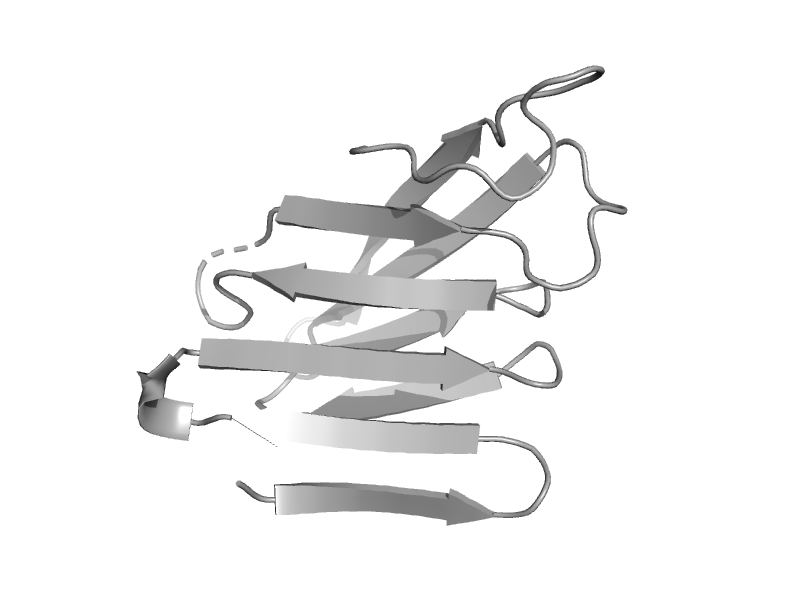 | view | |
| Hyaluronate lyase | Hyaluronate lyase-like, C-terminal domain | Pneumococcus (Streptococcus pneumoniae) [TaxId: 1313] | 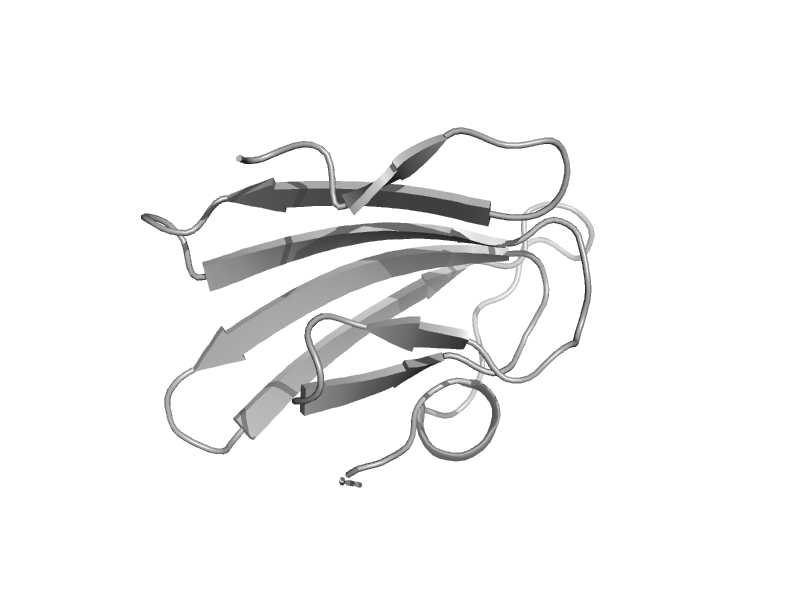 | view | |
| Hyaluronate lyase | Hyaluronate lyase-like, C-terminal domain | Streptococcus agalactiae [TaxId: 1311] |  | view | |
| Chondroitin ABC lyase I | Hyaluronate lyase-like, C-terminal domain | Proteus vulgaris [TaxId: 585] |  | view | |
| Chondroitinase AC | Hyaluronate lyase-like, C-terminal domain | Arthrobacter aurescens [TaxId: 43663] |  | view | |
| automated matches | automated matches | Bacillus sp. [TaxId: 84635] | 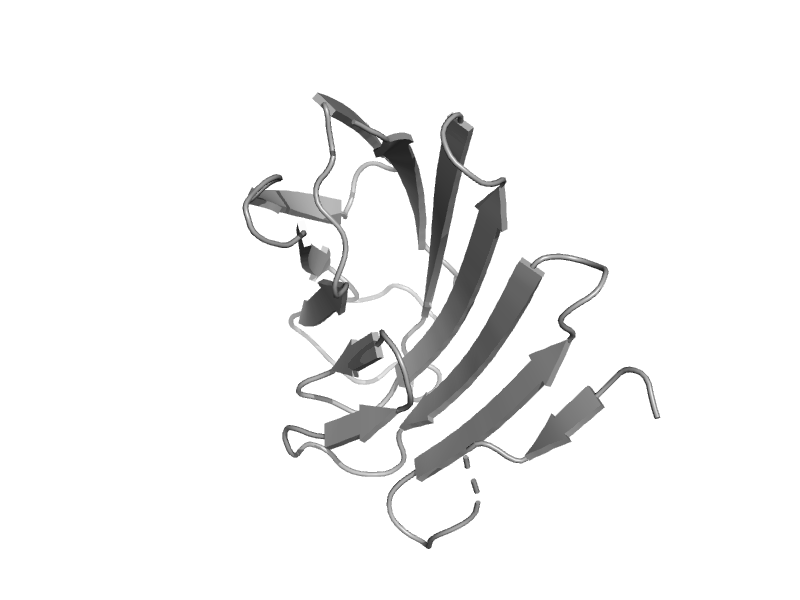 | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | P | K | V | L | A | N | T | N | Q | L | Q | A | V | Y | H | Q | Q | L | D | M | V | Q | A | I | F | Y | T | A | G | K | L | S | V | A | G | I | E | I | E | T | D | K | P | C | A | V | L | I | K | H | I | N | - | - | G | Q | V | I | W | A | A | D | P | L | Q | - | - | - | - | - | - | - | - | K | E | K | T | A | V | L | S | I | R | - | D | L | - | - | - | - | - | - | - | - | - | - | - | - | - | K | T | - | G | K | T | N | R | V | K | I | D | F | P | Q | Q | E | F | A | G | A | T | V | E | L | - | - | - | - | - | - |
| S | - | S | L | I | E | N | N | E | T | L | Q | S | V | Y | D | A | K | Q | G | V | W | G | I | V | K | Y | D | D | S | V | S | T | I | S | N | - | Q | F | Q | V | L | K | R | G | V | Y | T | I | R | K | E | G | - | D | E | Y | K | I | A | Y | Y | N | P | E | T | Q | E | S | A | P | D | Q | E | V | F | K | K | Q | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| E | V | E | L | L | E | N | S | S | K | Q | Q | V | I | Y | D | K | N | S | Q | T | W | A | V | I | K | H | D | N | Q | E | S | L | I | N | N | - | Q | F | K | M | N | K | A | G | L | Y | L | V | Q | K | V | G | - | N | D | Y | Q | N | V | Y | Y | Q | P | Q | T | M | T | K | T | D | Q | L | A | I | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | Y | Q | V | L | R | K | D | K | D | V | H | I | I | L | D | K | L | S | N | V | T | G | Y | A | F | Y | Q | P | A | S | I | E | D | - | K | W | I | K | K | V | N | K | P | A | I | V | M | T | H | R | Q | K | - | D | T | L | I | V | S | A | V | T | P | D | L | N | M | T | R | Q | K | A | A | T | P | V | T | I | N | V | T | I | N | - | G | K | W | Q | S | A | D | K | N | S | E | V | K | Y | Q | V | S | - | G | D | N | T | E | L | T | F | T | S | Y | - | - | - | - | F | G | I | P | Q | E | I | K | L | S | P | L | P |
| K | Y | S | V | I | R | N | D | A | T | A | Q | S | V | E | F | K | T | A | K | T | T | A | A | T | F | W | K | - | - | P | G | M | A | G | - | - | D | L | G | A | S | G | P | A | C | V | V | F | S | R | H | G | - | N | E | L | S | L | A | V | S | E | P | T | Q | K | A | A | - | - | - | - | - | - | - | - | G | L | T | L | T | L | P | E | G | T | W | S | S | V | L | E | G | A | - | G | T | L | G | T | D | A | D | G | R | S | T | L | T | L | D | T | T | - | - | G | L | S | G | K | T | K | L | I | K | L | K | R | - | - |
| P | A | I | E | I | V | N | T | S | G | V | Q | S | V | K | E | K | T | L | G | L | V | G | A | N | F | W | T | D | T | T | Q | T | A | D | - | - | L | I | T | S | N | K | K | A | S | V | M | T | R | E | I | A | D | E | R | L | E | A | S | V | S | D | P | T | Q | A | N | N | - | - | - | - | - | - | - | G | T | I | A | I | E | L | A | - | R | S | A | E | G | Y | S | A | D | P | G | I | T | V | T | Q | L | A | - | P | T | I | K | F | T | V | N | V | N | - | - | G | A | K | G | K | S | F | H | A | S | F | Q | L | G | - |
