solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1c3ga1 ( 180 ) etvqvnlpVsledLfvGkkksfkigrkGph---gasektqIdIq
d1c3ga2 ( 260 ) nfkrdgddLiYtLpLsfkeSllG--FsktiqTid-------grtlpLsrv
d1nlta1 ( 110 ) pqrgkdikheisAsleeLykG--rtaklaln--------rkileVhVe
d1nlta2 ( 258 ) sfkrdgddLvyeAeIdlltAiaG--gefaLehvs-------gdwlkvgIv
d2q2ga1 ( 4 ) rshevpLlVtleelylgkrkkikvtrkrfiehkvrneeniveVe
d2q2ga2 ( 87 ) rftrddchli-kvtIplvrAltg--ftCpvttld-------nrnlqipIk
d3agxa2 ( 246 ) ifkrdgsdViypArIslreAlcg--ctvnVpTld-------grtipvvFk
bbbbbbbb aaaaaa bbbbbb bbbbbb
60 70 80 90 100
d1c3ga1 ( 221 ) Lkpgwkagtkityknqgdynpq-tgrrktlqFvIqekshp
d1c3ga2 ( 301 ) q-pV-qpsqtstypgqGmptpknpsqrgnLiVkYkvdypislndaqkrai
d1nlta1 ( 222 ) --pgmkdgqrivfkgeAdqa-p-dvipgdVVFiVserphk
d1nlta2 ( 299 ) pgeViapgmrkviegkGmpipky-ggygnLiIkFtikdpe
d2q2ga1 ( 48 ) IkpgwkdgtkltysgeGdqesp-gtspgdLvLiIqtkthp
d2q2ga2 ( 128 ) e-iV-npktrkivpneG-piknqpgqkgdLILeFdIcfpksltpeqkkli
d3agxa2 ( 287 ) d-vI-rpgmrrkvpgeGlplpktpekrgdLiIeFeVifperipqtsrtvl
bbbb bbbbbbb
d1c3ga1
d1c3ga2 ( 349 ) d
d1nlta1
d1nlta2
d2q2ga1
d2q2ga2 ( 176 ) keal
d3agxa2 ( 335 ) eqvlpi
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| Heat shock protein 40 Sis1 | HSP40/DnaJ peptide-binding domain | Baker's yeast (Saccharomyces cerevisiae) [TaxId: 4932] | 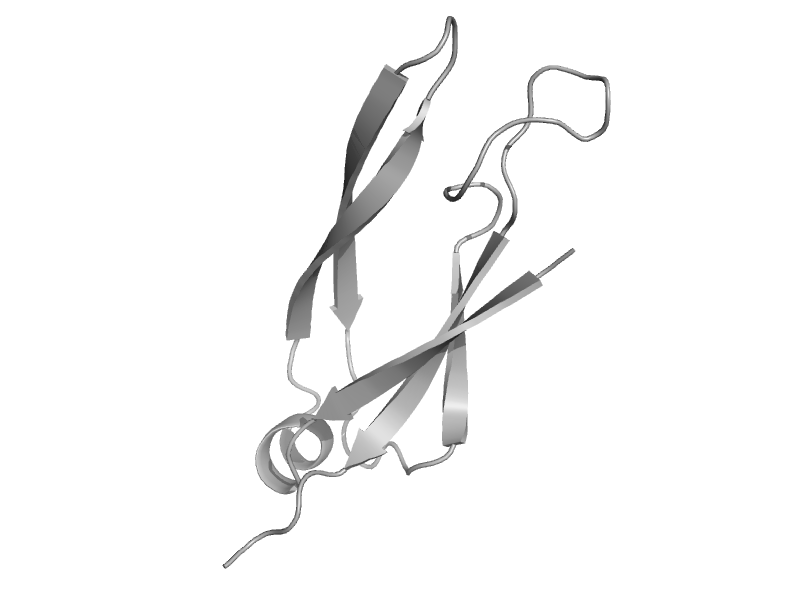 | view | |
| Heat shock protein 40 Sis1 | HSP40/DnaJ peptide-binding domain | Baker's yeast (Saccharomyces cerevisiae) [TaxId: 4932] | 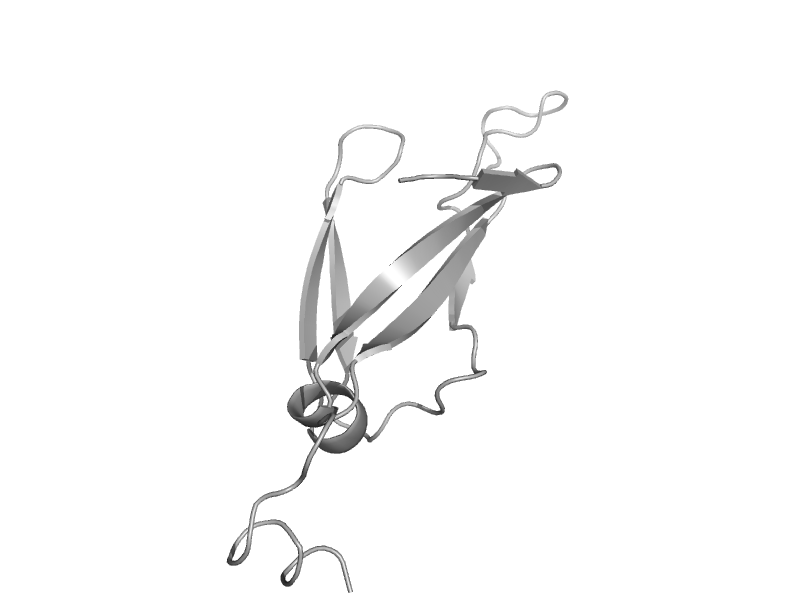 | view | |
| Mitochondrial protein import protein mas5 (Hsp40, Ydj1), C-terminal domain | HSP40/DnaJ peptide-binding domain | Baker's yeast (Saccharomyces cerevisiae) [TaxId: 4932] |  | view | |
| Mitochondrial protein import protein mas5 (Hsp40, Ydj1), C-terminal domain | HSP40/DnaJ peptide-binding domain | Baker's yeast (Saccharomyces cerevisiae) [TaxId: 4932] | 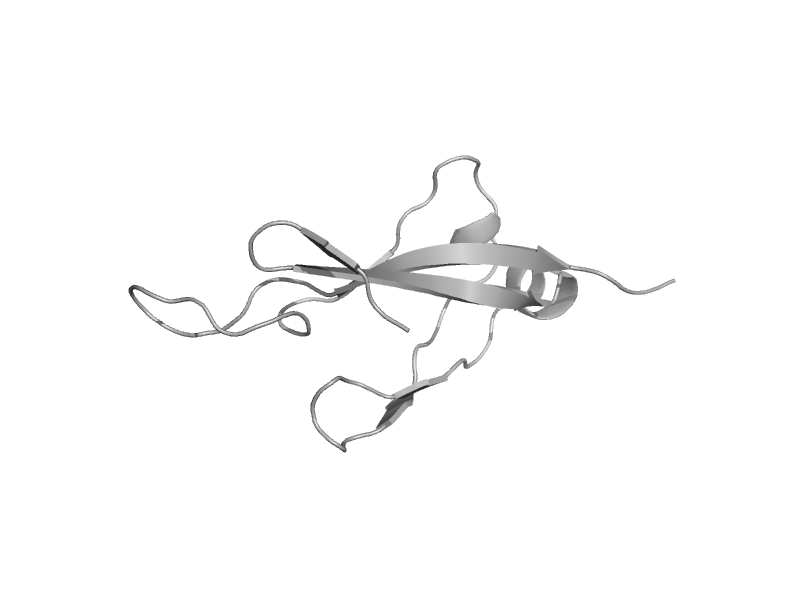 | view | |
| automated matches | automated matches | Cryptosporidium parvum [TaxId: 353152] | 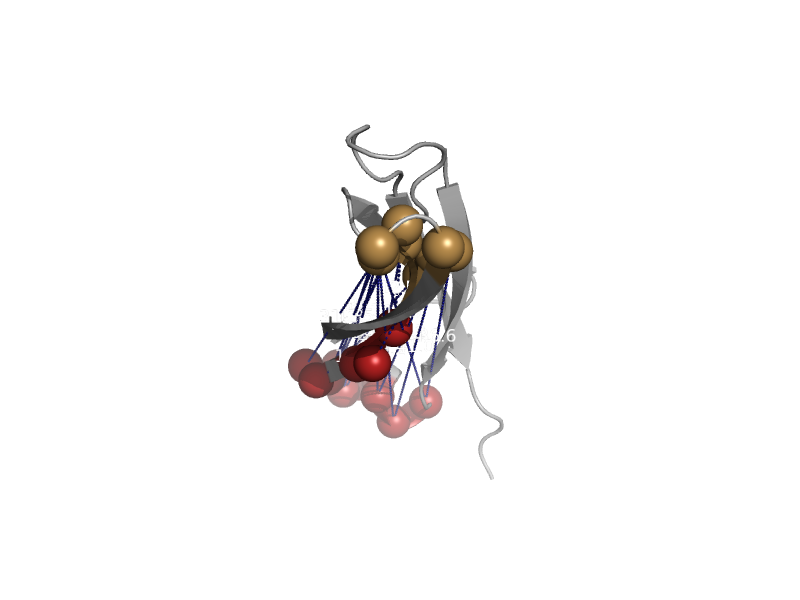 | view | |
| automated matches | automated matches | Cryptosporidium parvum [TaxId: 353152] | 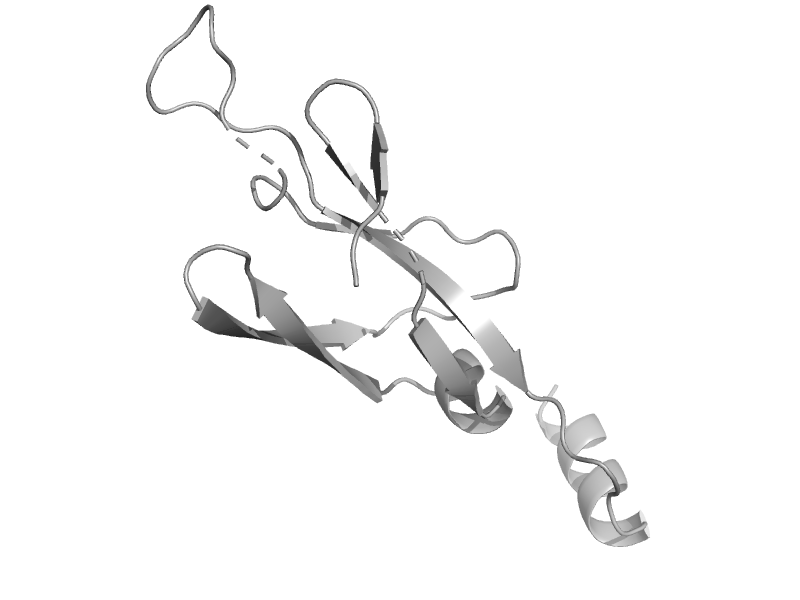 | view | |
| automated matches | automated matches | Human (Homo sapiens) [TaxId: 9606] | 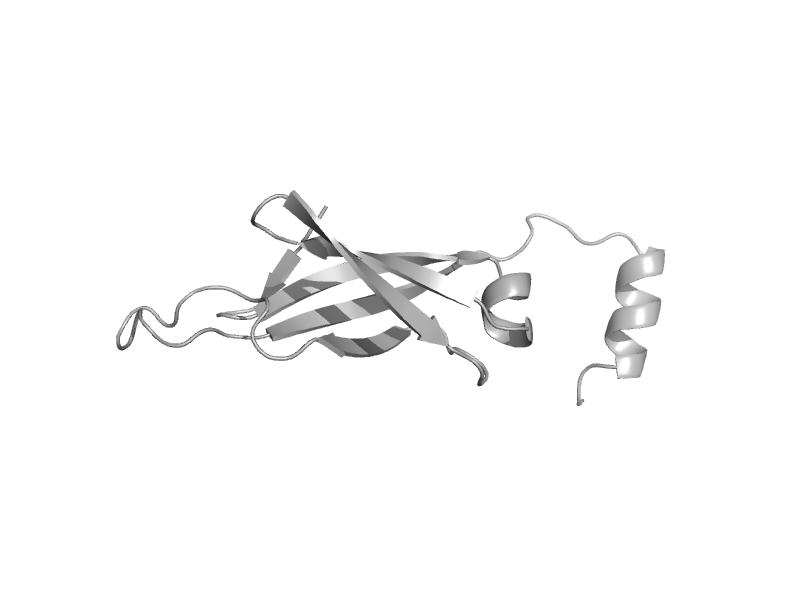 | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | - | - | - | - | - | E | T | V | Q | V | N | L | P | V | S | L | E | D | L | F | V | G | K | K | K | S | F | K | I | G | R | K | G | P | H | - | - | - | G | A | S | E | K | T | Q | I | D | I | Q | L | K | P | G | W | K | A | G | T | K | I | T | Y | K | N | Q | G | D | Y | N | P | Q | - | T | G | R | R | K | T | L | Q | F | V | I | Q | E | K | S | H | P | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| N | F | K | R | D | G | D | D | L | I | Y | T | L | P | L | S | F | K | E | S | L | L | G | - | - | F | S | K | T | I | Q | T | I | D | - | - | - | - | - | - | - | G | R | T | L | P | L | S | R | V | Q | - | P | V | - | Q | P | S | Q | T | S | T | Y | P | G | Q | G | M | P | T | P | K | N | P | S | Q | R | G | N | L | I | V | K | Y | K | V | D | Y | P | I | S | L | N | D | A | Q | K | R | A | I | D | - | - | - | - | - |
| - | - | P | Q | R | G | K | D | I | K | H | E | I | S | A | S | L | E | E | L | Y | K | G | - | - | R | T | A | K | L | A | L | N | - | - | - | - | - | - | - | - | R | K | I | L | E | V | H | V | E | - | - | P | G | M | K | D | G | Q | R | I | V | F | K | G | E | A | D | Q | A | - | P | - | D | V | I | P | G | D | V | V | F | I | V | S | E | R | P | H | K | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| S | F | K | R | D | G | D | D | L | V | Y | E | A | E | I | D | L | L | T | A | I | A | G | - | - | G | E | F | A | L | E | H | V | S | - | - | - | - | - | - | - | G | D | W | L | K | V | G | I | V | P | G | E | V | I | A | P | G | M | R | K | V | I | E | G | K | G | M | P | I | P | K | Y | - | G | G | Y | G | N | L | I | I | K | F | T | I | K | D | P | E | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | R | S | H | E | V | P | L | L | V | T | L | E | E | L | Y | L | G | K | R | K | K | I | K | V | T | R | K | R | F | I | E | H | K | V | R | N | E | E | N | I | V | E | V | E | I | K | P | G | W | K | D | G | T | K | L | T | Y | S | G | E | G | D | Q | E | S | P | - | G | T | S | P | G | D | L | V | L | I | I | Q | T | K | T | H | P | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| R | F | T | R | D | D | C | H | L | I | - | K | V | T | I | P | L | V | R | A | L | T | G | - | - | F | T | C | P | V | T | T | L | D | - | - | - | - | - | - | - | N | R | N | L | Q | I | P | I | K | E | - | I | V | - | N | P | K | T | R | K | I | V | P | N | E | G | - | P | I | K | N | Q | P | G | Q | K | G | D | L | I | L | E | F | D | I | C | F | P | K | S | L | T | P | E | Q | K | K | L | I | K | E | A | L | - | - |
| I | F | K | R | D | G | S | D | V | I | Y | P | A | R | I | S | L | R | E | A | L | C | G | - | - | C | T | V | N | V | P | T | L | D | - | - | - | - | - | - | - | G | R | T | I | P | V | V | F | K | D | - | V | I | - | R | P | G | M | R | R | K | V | P | G | E | G | L | P | L | P | K | T | P | E | K | R | G | D | L | I | I | E | F | E | V | I | F | P | E | R | I | P | Q | T | S | R | T | V | L | E | Q | V | L | P | I |
