solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1h8la1 ( 305 ) GIwGfVlDa------tdgrGilnAtIsVadin--hpvtTykdGdYwrl
d1uwya1 ( 297 ) GVkgqVfDq------n-gnplpnViVeVqdrkhiçpyrTnkyGeYyll
d5m0yb1 ( 8 ) kTtVsGyIsVdfdyppeseskiksgFnVkVagte--lstkTdekGyFeIs
bbbbbb bbbb bbbb
60 70 80 90 100
d1h8la1 ( 345 ) Lvq---gtykVtAsargydpvtktVeVdsk----ggvqvnFtLsrt
d1uwya1 ( 338 ) Llp---gsyiInVtVpghdphitkviIpeksqnfsalkkdIlLpfqgpsç
d5m0yb1 ( 56 ) gIpgdmreFtLeIskrnylkrnvtvngtg--------klvvstednpliL
bbbbbbb bbbbbb b
d1h8la1
d1uwya1 ( 394 ) pmiplyrnl
d5m0yb1 ( 98 ) wagdv
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| Carboxypeptidase D C-terminal domain | Carboxypeptidase regulatory domain | Crested duck (Lophonetta specularioides) [TaxId: 8836] | 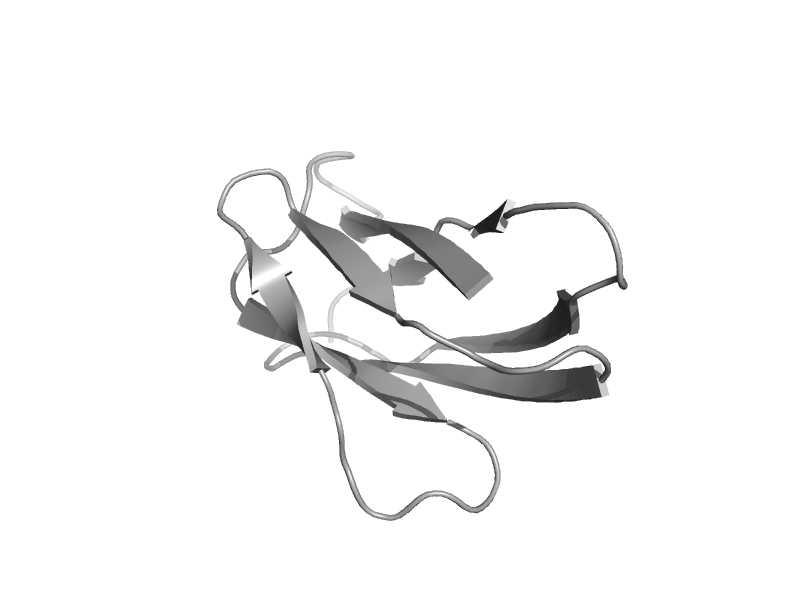 | view | |
| Carboxypeptidase M C-terminal domain | Carboxypeptidase regulatory domain | Human (Homo sapiens) [TaxId: 9606] | 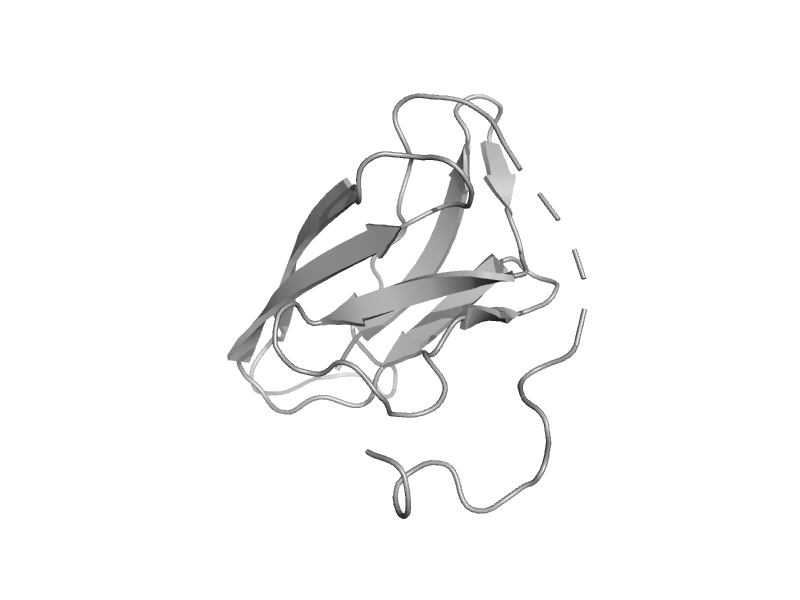 | view | |
| automated matches | automated matches | Clostridium thermocellum [TaxId: 203119] | 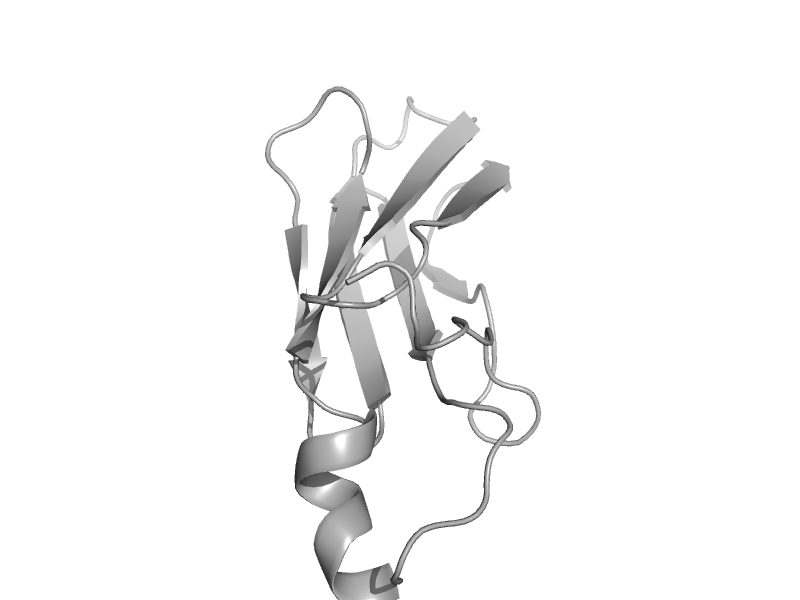 | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | - | G | I | W | G | F | V | L | D | A | - | - | - | - | - | - | T | D | G | R | G | I | L | N | A | T | I | S | V | A | D | I | N | - | - | H | P | V | T | T | Y | K | D | G | D | Y | W | R | L | L | V | Q | - | - | - | G | T | Y | K | V | T | A | S | A | R | G | Y | D | P | V | T | K | T | V | E | V | D | S | K | - | - | - | - | G | G | V | Q | V | N | F | T | L | S | R | T | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | G | V | K | G | Q | V | F | D | Q | - | - | - | - | - | - | N | - | G | N | P | L | P | N | V | I | V | E | V | Q | D | R | K | H | I | C | P | Y | R | T | N | K | Y | G | E | Y | Y | L | L | L | L | P | - | - | - | G | S | Y | I | I | N | V | T | V | P | G | H | D | P | H | I | T | K | V | I | I | P | E | K | S | Q | N | F | S | A | L | K | K | D | I | L | L | P | F | Q | G | P | S | C | P | M | I | P | L | Y | R | N | L |
| K | T | T | V | S | G | Y | I | S | V | D | F | D | Y | P | P | E | S | E | S | K | I | K | S | G | F | N | V | K | V | A | G | T | E | - | - | L | S | T | K | T | D | E | K | G | Y | F | E | I | S | G | I | P | G | D | M | R | E | F | T | L | E | I | S | K | R | N | Y | L | K | R | N | V | T | V | N | G | T | G | - | - | - | - | - | - | - | - | K | L | V | V | S | T | E | D | N | P | L | I | L | W | A | G | D | V | - | - | - | - |
| Member | Chain | UniProtKB | GO term |
|---|---|---|---|
| d1h8la1 | A | P83852 | GO:0004180 |
| d1h8la1 | A | P83852 | GO:0004181 |
| d1h8la1 | A | P83852 | GO:0005615 |
| d1h8la1 | A | P83852 | GO:0006508 |
| d1h8la1 | A | P83852 | GO:0006518 |
| d1h8la1 | A | P83852 | GO:0008237 |
| d1h8la1 | A | P83852 | GO:0008270 |
| d1h8la1 | A | P83852 | GO:0016020 |
| d1h8la1 | A | P83852 | GO:0016485 |
| d1h8la1 | A | P83852 | GO:0046872 |
| d1uwya1 | A | P14384 | GO:0004180 |
| d1uwya1 | A | P14384 | GO:0004181 |
| d1uwya1 | A | P14384 | GO:0005576 |
| d1uwya1 | A | P14384 | GO:0005615 |
| d1uwya1 | A | P14384 | GO:0005886 |
| d1uwya1 | A | P14384 | GO:0006508 |
| d1uwya1 | A | P14384 | GO:0006518 |
| d1uwya1 | A | P14384 | GO:0008237 |
| d1uwya1 | A | P14384 | GO:0008270 |
| d1uwya1 | A | P14384 | GO:0009653 |
| d1uwya1 | A | P14384 | GO:0009986 |
| d1uwya1 | A | P14384 | GO:0016485 |
| d1uwya1 | A | P14384 | GO:0046872 |
| d1uwya1 | A | P14384 | GO:0070062 |
| d1uwya1 | A | P14384 | GO:0098552 |
