solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1n67a2 ( 370 ) ekygkfynLSIKGtIdqidktnntYrQTIyVNPsGdnViaPvLTgNlk
d1zk5a- ( 1 ) aVsf-igs-teNdVgpsqgsysstnlpfvY-
d4x5pa- ( 1 ) FaÇktangtaipi-gGgsanvyvnla-pvvnvgqnlvvd-
d4z3ga1 ( 0 ) awnnivFYSlgdvnsyqggnvvitqrpqFitsWrp--giAt-vtwnqçng
d5ihwa2 ( 424 ) qdpmvhgdSNIQSifTkldedkqtIeQQIYVNplkksAtnTkVdIAGS
bbb bb bbbbbb bb bbb
60 70 80 90 100
d1n67a2 ( 418 ) p--------ntdSNAlIdqqnTsIkVykVdna-ad-LsesyfV-npenfe
d1zk5a- ( 34 ) --tghnIGyq-nanVWIsggfÇVGLdGkVdlpv--vgsld----------
d4x5pa- ( 38 ) --lstqifÇh-ndypeti-tDyvtlqrGsAy-------------------
d4z3ga1 ( 47 ) pe----fAdgswaYyrey-iAWvvFpkkvmtkngyplf--ievhnkgsWs
d5ihwa2 ( 472 ) qVddyGniklgngsTiIdq-nTeIkVykVn-s-dqqLpqsNrIydfsqYe
b bbbbbbb
110 120 130 140 150
d1n67a2 ( 457 ) dvtnsV--nitfp----npnQYkVeFntpddqIt---tPYiVvVnGhIdp
d1zk5a- ( 70 ) -------gqsIYGLteeVGLlIwMGdtnysrGtaMsgnswenVfsGwçVg
d4x5pa- ( 65 ) ------------ggvlsnFsgtvkYsg-----ssypFpttsetprvvYns
d4z3ga1 ( 90 ) eentgdn------------------------------dSyFFlkgykwdq
d5ihwa2 ( 519 ) dVtsqFdnkksfs-----nnvAtLdFg----dIn---sAyIIkVvSkYtp
bbbbb b bbbbbbbb
160 170 180 190 200
d1n67a2 ( 498 ) ns--------------k--gdLALrSTLYGynsn------iiWrSMSWDN
d1zk5a- ( 113 ) n---------------yvstQGLSVhVrPVilnssaqysVqk-tsIGsIr
d4x5pa- ( 98 ) r-----------------tdkpWpvalyltPvssAggvAikagsliAvli
d4z3ga1 ( 110 ) rafdtanlçqkpgettrltekfddiiFkvalpadlplgdysvtipYtSGi
d5ihwa2 ( 557 ) ts--------------dgeldIAQGTsMrTtdky------gyynyAGysn
b bbbbbbb bbbbbbb
210 220 230 240
d1n67a2 ( 526 ) evafnngsgsgdgidkpvvPeqPdepgeiepiPe
d1zk5a- ( 149 ) Mrpy-ngssag-----------svqttVnFsL-npFtLnd
d4x5pa- ( 131 ) lrqtnnynsdd------------fqFvWniyAnndvvvpt
d4z3ga1 ( 160 ) qrHFAsyl---gArfkipynvAktlpreneMlFlFknigg
d5ihwa2 ( 587 ) fivts
bbb
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| Clumping factor A | Fibrinogen-binding domain | Staphylococcus aureus [TaxId: 1280] | 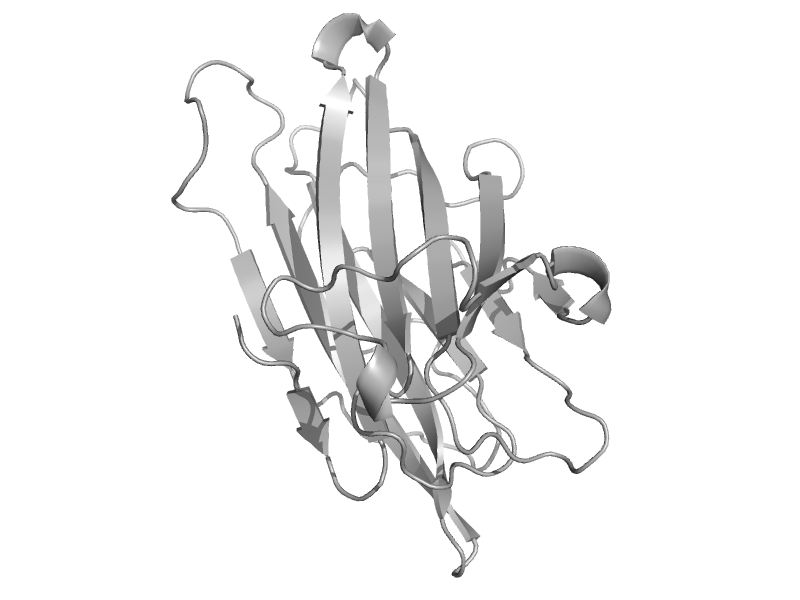 | view | |
| automated matches | F17c-type adhesin | Escherichia coli [TaxId: 562] | 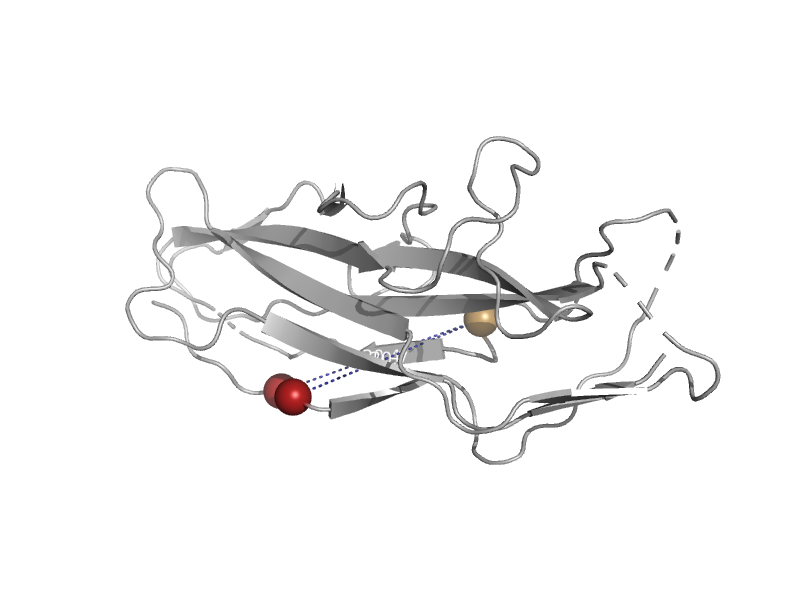 | view | |
| Mannose-specific adhesin FimH, N-terminal domain | Pilus subunits | Escherichia coli [TaxId: 83333] | 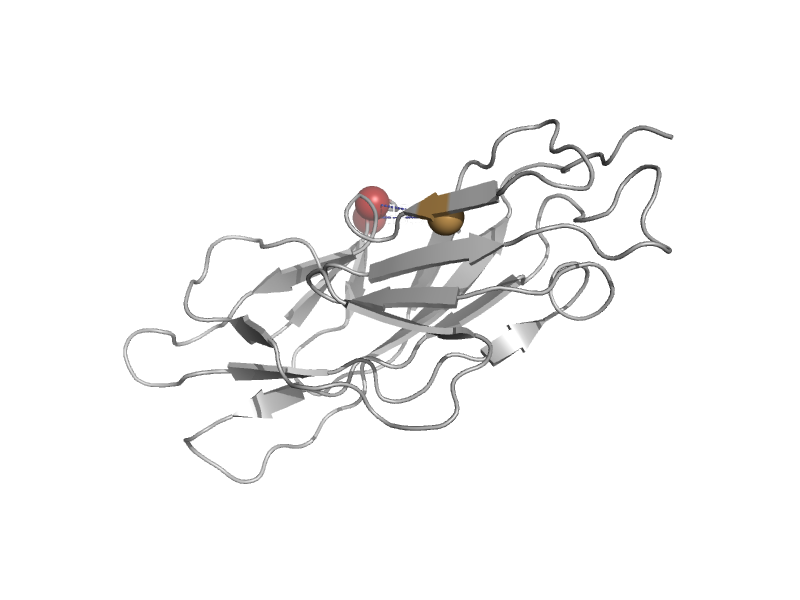 | view | |
| automated matches | PapG adhesin receptor-binding domain | Escherichia coli [TaxId: 562] | 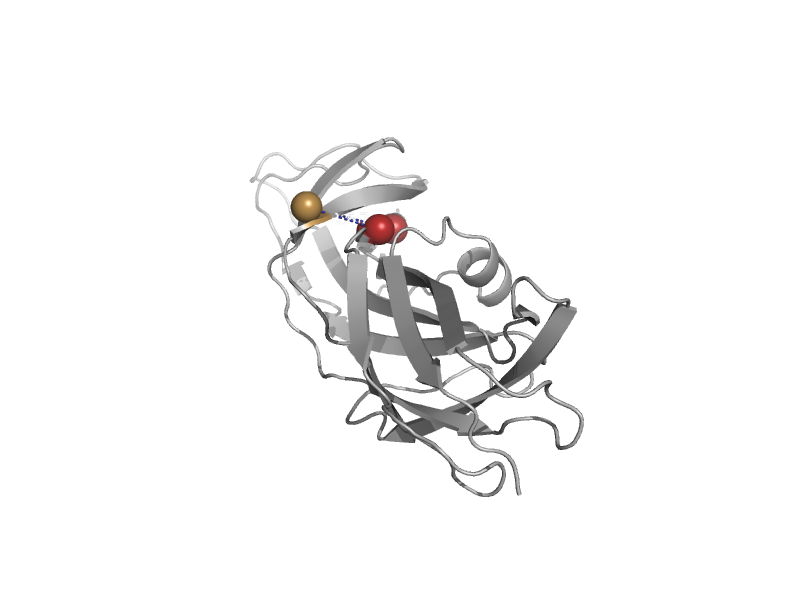 | view | |
| automated matches | automated matches | Staphylococcus aureus [TaxId: 282459] |  | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | 150 | 160 | 170 | 180 | 190 | 200 | 210 | 220 | 230 | 240 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | - | E | K | Y | G | K | F | Y | N | L | S | I | K | G | T | I | D | Q | I | D | K | T | N | N | T | Y | R | Q | T | I | Y | V | N | P | S | G | D | N | V | I | A | P | V | L | T | G | N | L | K | P | - | - | - | - | - | - | - | - | N | T | D | S | N | A | L | I | D | Q | Q | N | T | S | I | K | V | Y | K | V | D | N | A | - | A | D | - | L | S | E | S | Y | F | V | - | N | P | E | N | F | E | D | V | T | N | S | V | - | - | N | I | T | F | P | - | - | - | - | N | P | N | Q | Y | K | V | E | F | N | T | P | D | D | Q | I | T | - | - | - | T | P | Y | I | V | V | V | N | G | H | I | D | P | N | S | - | - | - | - | - | - | - | - | - | - | - | - | - | - | K | - | - | G | D | L | A | L | R | S | T | L | Y | G | Y | N | S | N | - | - | - | - | - | - | I | I | W | R | S | M | S | W | D | N | E | V | A | F | N | N | G | S | G | S | G | D | G | I | D | K | P | V | V | P | E | Q | P | D | E | P | G | E | I | E | P | I | P | E | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | A | V | S | F | - | I | G | S | - | T | E | N | D | V | G | P | S | Q | G | S | Y | S | S | T | N | L | P | F | V | Y | - | - | - | T | G | H | N | I | G | Y | Q | - | N | A | N | V | W | I | S | G | G | F | C | V | G | L | D | G | K | V | D | L | P | V | - | - | V | G | S | L | D | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | Q | S | I | Y | G | L | T | E | E | V | G | L | L | I | W | M | G | D | T | N | Y | S | R | G | T | A | M | S | G | N | S | W | E | N | V | F | S | G | W | C | V | G | N | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Y | V | S | T | Q | G | L | S | V | H | V | R | P | V | I | L | N | S | S | A | Q | Y | S | V | Q | K | - | T | S | I | G | S | I | R | M | R | P | Y | - | N | G | S | S | A | G | - | - | - | - | - | - | - | - | - | - | - | S | V | Q | T | T | V | N | F | S | L | - | N | P | F | T | L | N | D |
| - | - | - | - | - | - | - | - | - | - | F | A | C | K | T | A | N | G | T | A | I | P | I | - | G | G | G | S | A | N | V | Y | V | N | L | A | - | P | V | V | N | V | G | Q | N | L | V | V | D | - | - | - | L | S | T | Q | I | F | C | H | - | N | D | Y | P | E | T | I | - | T | D | Y | V | T | L | Q | R | G | S | A | Y | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | G | V | L | S | N | F | S | G | T | V | K | Y | S | G | - | - | - | - | - | S | S | Y | P | F | P | T | T | S | E | T | P | R | V | V | Y | N | S | R | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | T | D | K | P | W | P | V | A | L | Y | L | T | P | V | S | S | A | G | G | V | A | I | K | A | G | S | L | I | A | V | L | I | L | R | Q | T | N | N | Y | N | S | D | D | - | - | - | - | - | - | - | - | - | - | - | - | F | Q | F | V | W | N | I | Y | A | N | N | D | V | V | V | P | T |
| A | W | N | N | I | V | F | Y | S | L | G | D | V | N | S | Y | Q | G | G | N | V | V | I | T | Q | R | P | Q | F | I | T | S | W | R | P | - | - | G | I | A | T | - | V | T | W | N | Q | C | N | G | P | E | - | - | - | - | F | A | D | G | S | W | A | Y | Y | R | E | Y | - | I | A | W | V | V | F | P | K | K | V | M | T | K | N | G | Y | P | L | F | - | - | I | E | V | H | N | K | G | S | W | S | E | E | N | T | G | D | N | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | D | S | Y | F | F | L | K | G | Y | K | W | D | Q | R | A | F | D | T | A | N | L | C | Q | K | P | G | E | T | T | R | L | T | E | K | F | D | D | I | I | F | K | V | A | L | P | A | D | L | P | L | G | D | Y | S | V | T | I | P | Y | T | S | G | I | Q | R | H | F | A | S | Y | L | - | - | - | G | A | R | F | K | I | P | Y | N | V | A | K | T | L | P | R | E | N | E | M | L | F | L | F | K | N | I | G | G |
| - | - | Q | D | P | M | V | H | G | D | S | N | I | Q | S | I | F | T | K | L | D | E | D | K | Q | T | I | E | Q | Q | I | Y | V | N | P | L | K | K | S | A | T | N | T | K | V | D | I | A | G | S | Q | V | D | D | Y | G | N | I | K | L | G | N | G | S | T | I | I | D | Q | - | N | T | E | I | K | V | Y | K | V | N | - | S | - | D | Q | Q | L | P | Q | S | N | R | I | Y | D | F | S | Q | Y | E | D | V | T | S | Q | F | D | N | K | K | S | F | S | - | - | - | - | - | N | N | V | A | T | L | D | F | G | - | - | - | - | D | I | N | - | - | - | S | A | Y | I | I | K | V | V | S | K | Y | T | P | T | S | - | - | - | - | - | - | - | - | - | - | - | - | - | - | D | G | E | L | D | I | A | Q | G | T | S | M | R | T | T | D | K | Y | - | - | - | - | - | - | G | Y | Y | N | Y | A | G | Y | S | N | F | I | V | T | S | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Member | Chain | UniProtKB | GO term |
|---|---|---|---|
| d1n67a2 | A | Q53653 | GO:0005618 |
| d1n67a2 | A | Q53653 | GO:0007155 |
| d1n67a2 | A | Q53653 | GO:0005618 |
| d1n67a2 | A | Q53653 | GO:0007155 |
| d1n67a2 | A | Q53653 | GO:0005618 |
| d1n67a2 | A | Q53653 | GO:0007155 |
| d1n67a2 | A | Q53653 | GO:0005618 |
| d1n67a2 | A | Q53653 | GO:0007155 |
