solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1iqra1 ( 172 ) lplpepgeeaAlagLraFLe--akLpry-aeerdrLdg
d1np7a1 ( 205 ) dhrsVlaFqggetaGlarLqdYFwhgdrLkdY-ketrngMvg
d1owla1 ( 205 ) pvepgetaAiarLqeFCd--raIady-dpqrnfPae
d1u3da1 ( 198 ) eddsekgsnallarawspgwsnGdkaLttFIn--gpLley-sknrrkAds
d2wq7a2 ( 216 ) gpnkFpggeteAlrrMeesLkdeiwVArFekpntaPNsl
aaaaaaaaaaaa 333
60 70 80 90 100
d1iqra1 ( 207 ) e-ggsrLspyfAlgvLsprlAaweAerrg-----------geGArkWvae
d1np7a1 ( 246 ) adyssKFspWlAlgcLsprfIyqeVkrYEqer------vsndSThwlife
d1owla1 ( 238 ) a-gtsgLspAlkfgaIgirqAwqaAsaahals---rsdearnSIrvWQqe
d1u3da1 ( 245 ) a-ttsfLsphlhfgeVsvrkVfhlVrikqvawanegneaGeeSVnlFlks
d2wq7a2 ( 255 ) epsttVLsPYlkfgcLsarlFnqkLkeIikrqp-----khsqppvsliGq
aaaaa aaaaaaaaaaaaaa aaaaaaaaa
110 120 130 140 150
d1iqra1 ( 245 ) llwrdfSyhLlyhfpwMaerPldprFqaFpwqedealFqaWyeGkTgVPL
d1np7a1 ( 290 ) llwrdfFrfvaqkygnkLfnrgGLlnknfpWqedqvrfelWrsGqTGYPL
d1owla1 ( 284 ) lawrefyqhAlyhfpsLadGPyrslWqqFpWenreaLftaWtqAqTGyPI
d1u3da1 ( 294 ) iglreySryisfnhpySherpllghLkfFpwavdenyfkaWrqGrTGYPL
d2wq7a2 ( 300 ) lmwrefYytvAaaepnFdrmlgNvyCmqIpWqehpdhleaWthGrTGYPF
aaaaaaaaaaaa aaaaaaaa aa
160 170 180 190 200
d1iqra1 ( 295 ) VDAAMreLhatGfLsnrArmNAAQFAVK-HLLLpWkrCEeaFrhlLlDgd
d1np7a1 ( 340 ) VDANMrELnltgFMSnrGrqNVASFLCK-NLgIDWrwGAewFescLiDyd
d1owla1 ( 334 ) VDAAMrqLtetgwMHnrCrmIVASFLTK-DLIIdWrrGeqfFmqhLvdGd
d1u3da1 ( 344 ) VDAGMrELwatgwLhdrIrvvVSSFFVK-VLQLpWrwGMkyFwdtLldad
d2wq7a2 ( 350 ) IDAIMrqLrqEgwIHhlArhAVACFLTRGDLWIsWeeGqrvFeqlLldqd
aaaaaaaaaaa aaaaaaaaaaaaa aaaaaaaaaaa
210 220 230 240 250
d1iqra1 ( 344 ) ravnLqgWQwaGGLgVdaapyfrVfnpvlQGerhdpegrWLkrwApeYp-
d1np7a1 ( 389 ) vcsnwGnWNytAgIG-ndardfryfnIpkQSqqydpqGtYLrhWLpeLkn
d1owla1 ( 383 ) laanNggWQwSAsSGmdpkp-lrIfnPasQAkkfDatAtYIkrwLpeLrh
d1u3da1 ( 393 ) lesdAlgWQyITGTLpdSrefdrIdnPqfeGykFDpnGeYVrrwLpeLsr
d2wq7a2 ( 400 ) wAlnAGnWMwLSASA-ffhqyfrVysPvafGkktdpqGhYIrkYVpeLsk
aaaaaaaaaaa aaaaaaa aaaaaa 333
260 270 280 290 300
d1iqra1 ( 393 ) -----sYapk-------------------dpvvdleeArrrYlrlArd
d1np7a1 ( 438 ) LpgdkIHqPwlLsateqkqwgVqLgvdYprpcvnfhqsVearrkie
d1owla1 ( 432 ) VhpkdLisge-Itpierr--------gYpapivnhnlrqkqFkalynqlk
d1u3da1 ( 443 ) LptdwIHhPwnApesvLqaAgIelgsnYplpivgldeAkarLheAlsqMw
d2wq7a2 ( 449 ) YpagcIYePwkAslvdqraygCvlgtdYphrivkhevvhkeNikrMgaAy
aaaa aaaaaaaaaaaaa
310
d1iqra1
d1np7a1
d1owla1 ( 473 ) aai
d1u3da1 ( 493 ) qleaa
d2wq7a2 ( 499 ) kvnrevrtgke
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| C-terminal domain of DNA photolyase | Cryptochrome/photolyase FAD-binding domain | Thermus thermophilus [TaxId: 274] | 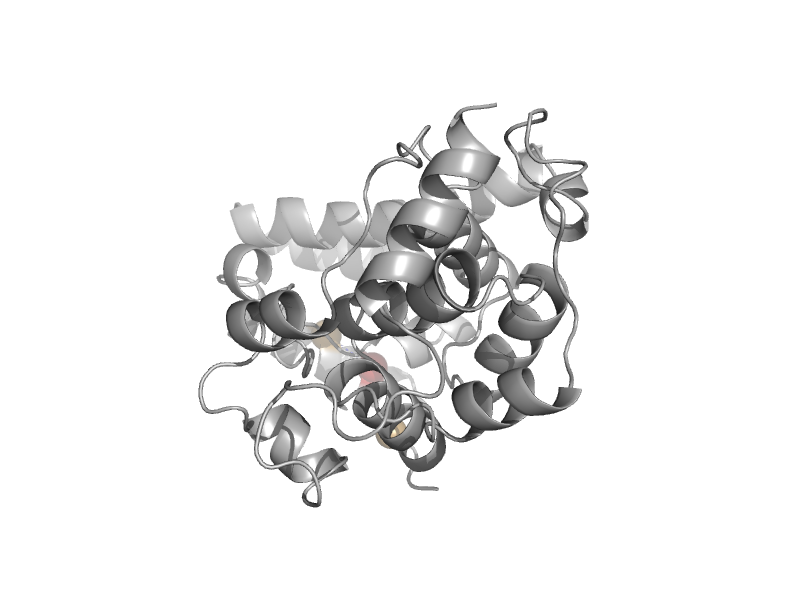 | view | |
| Cryptochrome C-terminal domain | Cryptochrome/photolyase FAD-binding domain | Synechocystis sp. PCC 6803 [TaxId: 1148] | 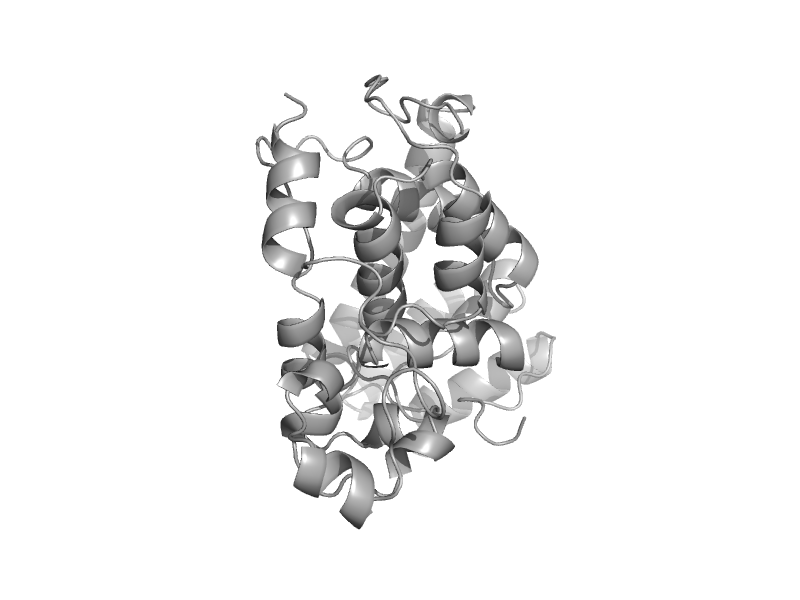 | view | |
| C-terminal domain of DNA photolyase | Cryptochrome/photolyase FAD-binding domain | Synechococcus elongatus [TaxId: 32046] | 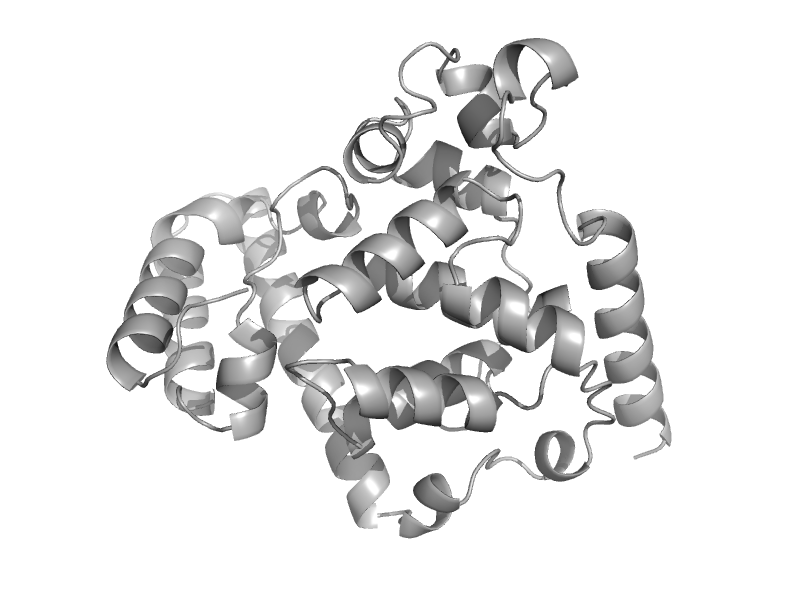 | view | |
| Cryptochrome C-terminal domain | Cryptochrome/photolyase FAD-binding domain | Thale cress (Arabidopsis thaliana) [TaxId: 3702] | 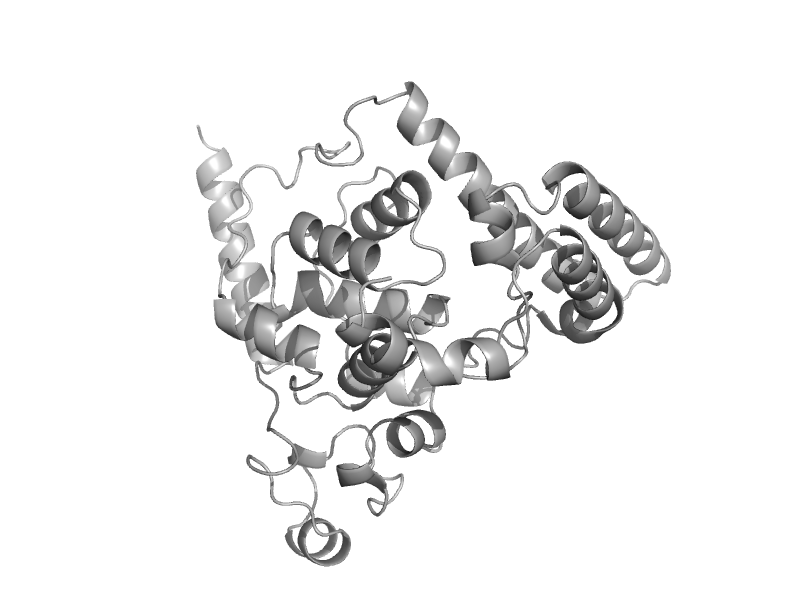 | view | |
| automated matches | automated matches | Fruit fly (Drosophila melanogaster) [TaxId: 7227] |  | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | 150 | 160 | 170 | 180 | 190 | 200 | 210 | 220 | 230 | 240 | 250 | 260 | 270 | 280 | 290 | 300 | 310 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | - | - | - | - | - | - | - | - | - | - | - | L | P | L | P | E | P | G | E | E | A | A | L | A | G | L | R | A | F | L | E | - | - | A | K | L | P | R | Y | - | A | E | E | R | D | R | L | D | G | E | - | G | G | S | R | L | S | P | Y | F | A | L | G | V | L | S | P | R | L | A | A | W | E | A | E | R | R | G | - | - | - | - | - | - | - | - | - | - | - | G | E | G | A | R | K | W | V | A | E | L | L | W | R | D | F | S | Y | H | L | L | Y | H | F | P | W | M | A | E | R | P | L | D | P | R | F | Q | A | F | P | W | Q | E | D | E | A | L | F | Q | A | W | Y | E | G | K | T | G | V | P | L | V | D | A | A | M | R | E | L | H | A | T | G | F | L | S | N | R | A | R | M | N | A | A | Q | F | A | V | K | - | H | L | L | L | P | W | K | R | C | E | E | A | F | R | H | L | L | L | D | G | D | R | A | V | N | L | Q | G | W | Q | W | A | G | G | L | G | V | D | A | A | P | Y | F | R | V | F | N | P | V | L | Q | G | E | R | H | D | P | E | G | R | W | L | K | R | W | A | P | E | Y | P | - | - | - | - | - | - | S | Y | A | P | K | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | D | P | V | V | D | L | E | E | A | R | R | R | Y | L | R | L | A | R | D | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | D | H | R | S | V | L | A | F | Q | G | G | E | T | A | G | L | A | R | L | Q | D | Y | F | W | H | G | D | R | L | K | D | Y | - | K | E | T | R | N | G | M | V | G | A | D | Y | S | S | K | F | S | P | W | L | A | L | G | C | L | S | P | R | F | I | Y | Q | E | V | K | R | Y | E | Q | E | R | - | - | - | - | - | - | V | S | N | D | S | T | H | W | L | I | F | E | L | L | W | R | D | F | F | R | F | V | A | Q | K | Y | G | N | K | L | F | N | R | G | G | L | L | N | K | N | F | P | W | Q | E | D | Q | V | R | F | E | L | W | R | S | G | Q | T | G | Y | P | L | V | D | A | N | M | R | E | L | N | L | T | G | F | M | S | N | R | G | R | Q | N | V | A | S | F | L | C | K | - | N | L | G | I | D | W | R | W | G | A | E | W | F | E | S | C | L | I | D | Y | D | V | C | S | N | W | G | N | W | N | Y | T | A | G | I | G | - | N | D | A | R | D | F | R | Y | F | N | I | P | K | Q | S | Q | Q | Y | D | P | Q | G | T | Y | L | R | H | W | L | P | E | L | K | N | L | P | G | D | K | I | H | Q | P | W | L | L | S | A | T | E | Q | K | Q | W | G | V | Q | L | G | V | D | Y | P | R | P | C | V | N | F | H | Q | S | V | E | A | R | R | K | I | E | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | P | V | E | P | G | E | T | A | A | I | A | R | L | Q | E | F | C | D | - | - | R | A | I | A | D | Y | - | D | P | Q | R | N | F | P | A | E | A | - | G | T | S | G | L | S | P | A | L | K | F | G | A | I | G | I | R | Q | A | W | Q | A | A | S | A | A | H | A | L | S | - | - | - | R | S | D | E | A | R | N | S | I | R | V | W | Q | Q | E | L | A | W | R | E | F | Y | Q | H | A | L | Y | H | F | P | S | L | A | D | G | P | Y | R | S | L | W | Q | Q | F | P | W | E | N | R | E | A | L | F | T | A | W | T | Q | A | Q | T | G | Y | P | I | V | D | A | A | M | R | Q | L | T | E | T | G | W | M | H | N | R | C | R | M | I | V | A | S | F | L | T | K | - | D | L | I | I | D | W | R | R | G | E | Q | F | F | M | Q | H | L | V | D | G | D | L | A | A | N | N | G | G | W | Q | W | S | A | S | S | G | M | D | P | K | P | - | L | R | I | F | N | P | A | S | Q | A | K | K | F | D | A | T | A | T | Y | I | K | R | W | L | P | E | L | R | H | V | H | P | K | D | L | I | S | G | E | - | I | T | P | I | E | R | R | - | - | - | - | - | - | - | - | G | Y | P | A | P | I | V | N | H | N | L | R | Q | K | Q | F | K | A | L | Y | N | Q | L | K | A | A | I | - | - | - | - | - | - | - | - |
| E | D | D | S | E | K | G | S | N | A | L | L | A | R | A | W | S | P | G | W | S | N | G | D | K | A | L | T | T | F | I | N | - | - | G | P | L | L | E | Y | - | S | K | N | R | R | K | A | D | S | A | - | T | T | S | F | L | S | P | H | L | H | F | G | E | V | S | V | R | K | V | F | H | L | V | R | I | K | Q | V | A | W | A | N | E | G | N | E | A | G | E | E | S | V | N | L | F | L | K | S | I | G | L | R | E | Y | S | R | Y | I | S | F | N | H | P | Y | S | H | E | R | P | L | L | G | H | L | K | F | F | P | W | A | V | D | E | N | Y | F | K | A | W | R | Q | G | R | T | G | Y | P | L | V | D | A | G | M | R | E | L | W | A | T | G | W | L | H | D | R | I | R | V | V | V | S | S | F | F | V | K | - | V | L | Q | L | P | W | R | W | G | M | K | Y | F | W | D | T | L | L | D | A | D | L | E | S | D | A | L | G | W | Q | Y | I | T | G | T | L | P | D | S | R | E | F | D | R | I | D | N | P | Q | F | E | G | Y | K | F | D | P | N | G | E | Y | V | R | R | W | L | P | E | L | S | R | L | P | T | D | W | I | H | H | P | W | N | A | P | E | S | V | L | Q | A | A | G | I | E | L | G | S | N | Y | P | L | P | I | V | G | L | D | E | A | K | A | R | L | H | E | A | L | S | Q | M | W | Q | L | E | A | A | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | G | P | N | K | F | P | G | G | E | T | E | A | L | R | R | M | E | E | S | L | K | D | E | I | W | V | A | R | F | E | K | P | N | T | A | P | N | S | L | E | P | S | T | T | V | L | S | P | Y | L | K | F | G | C | L | S | A | R | L | F | N | Q | K | L | K | E | I | I | K | R | Q | P | - | - | - | - | - | K | H | S | Q | P | P | V | S | L | I | G | Q | L | M | W | R | E | F | Y | Y | T | V | A | A | A | E | P | N | F | D | R | M | L | G | N | V | Y | C | M | Q | I | P | W | Q | E | H | P | D | H | L | E | A | W | T | H | G | R | T | G | Y | P | F | I | D | A | I | M | R | Q | L | R | Q | E | G | W | I | H | H | L | A | R | H | A | V | A | C | F | L | T | R | G | D | L | W | I | S | W | E | E | G | Q | R | V | F | E | Q | L | L | L | D | Q | D | W | A | L | N | A | G | N | W | M | W | L | S | A | S | A | - | F | F | H | Q | Y | F | R | V | Y | S | P | V | A | F | G | K | K | T | D | P | Q | G | H | Y | I | R | K | Y | V | P | E | L | S | K | Y | P | A | G | C | I | Y | E | P | W | K | A | S | L | V | D | Q | R | A | Y | G | C | V | L | G | T | D | Y | P | H | R | I | V | K | H | E | V | V | H | K | E | N | I | K | R | M | G | A | A | Y | K | V | N | R | E | V | R | T | G | K | E |
