solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1hc1a2 ( 136 )
d1js8a1 (2503 ) aiiR
d1llaa2 ( 110 )
d3awua1 ( 2 ) tvR
d4z0ya- ( 1 ) apitAPditsiÇkdAssgignqegAirtrkÇçppslgkikdffpndvrmR
60 70 80 90 100
d1hc1a2 ( 136 ) plyqit-phmFtnsevidkaysakmtqkpgtfnvsfknreqrVayfged
d1js8a1 (2507 ) knVnsLtpsDikeLrdAMakVqad--t------------------sdnGY
d1llaa2 ( 110 ) pVqeif-pDkFipsaaineafkki------lvt-gildpeyrLayyRed
d3awua1 ( 5 ) knQatltadekrrFvaAVleLKrs--g---------------------rY
d4z0ya- ( 55 ) WpAhg-tkkqvddYrrAiaaMralpdd------------------dPrSF
aaaaaaaaa
110 120 130 140 150
d1hc1a2 ( 188 ) igmNihHVtwHmdfPfwwedsygyhldrK--GelFfwvHhqLTarFdfeR
d1js8a1 (2537 ) qkIAsYHGipl-sÇhy---engtayaÇCqhgmVTFPNWHRLLTKQMEdAL
d1llaa2 ( 167 ) vgiNahHwhwhlvypstwnpyfgkkkdrK--gelFyymHqqMCarYdcer
d3awua1 ( 32 ) defVrtHnefimsDtd---s----gerTGhrSPSFLPWhRrFLLdFEqAL
d4z0ya- ( 87 ) vsqAkiHÇAYCngGYtqvdsgfpdidiqiHnSWlFFPFHRWYlYFYEril
aaaaaaaaaaa aaaaaaaaaaaaaaaa
160 170 180 190 200
d1hc1a2 ( 236 ) lsnwld---pVdel-hWd------riIreGfaPlTsyky-ggeFPvrp--
d1js8a1 (2583 ) vakg--ShvGIPYWdWTttfanLpvLVteek--------------dNsFh
d1llaa2 ( 216 ) lsngmh---rMlpfnnFd------epL-agyaPhLthvasgkyySpRp--
d3awua1 ( 75 ) qsvd--ssVtLPYWdWSadrtvrasLW-apdFLGgtGrstdgrVmdGpFA
d4z0ya- ( 137 ) GslidepnFAlPYWKWdepkGMpisiFlg-------dasnplydqyRDan
aa
210 220 230 240 250
d1hc1a2 ( 273 ) --------------dnihFedVdgvahvhdLeitesrIheAIdh--GyIt
d1js8a1 (2617 ) ----hAhIdvan---tdTtrspraq-LfsfFyrqIAlALeq-t-------
d1llaa2 ( 254 ) --------------dglkLrdL-gdIeiseMvrmrerIldSIhl--gyVi
d3awua1 ( 122 ) astgnWpInvrvdsrtyLrRsLggsvaeLPtraeVesVLai-saYDlppy
d4z0ya- ( 181 ) Hiedrvdldydgdkd---ipdqqvacnlstvyrDlvrnGvdptsFFGgkY
aaaaaaaaaa
260 270 280 290 300
d1hc1a2 ( 307 ) dsdghtidIrqpkGielLGdIIESSkySsnvqyYgsLHntAhvMLgrQgd
d1js8a1 (2659 ) dFç-------------dFEiqFEi------------gHnaIhswVGGs--
d1llaa2 ( 287 ) sedgshktLdelhGtdiLGaLVESSyeSvnheyYgnLHnwGhvTMArIhD
d3awua1 ( 171 ) nsaSe-----------GFRNhLEGwrg-------vnLhnrVhvWVg----
d4z0ya- ( 231 ) vAgdspvan-----gdpsvGsvea------------gStAvhrWvGdptq
aaaaaa aaaaaaaa
310 320 330 340 350
d1hc1a2 ( 357 ) phgkfnlppgVMehfeTATrDpSFFrLHkYMdnIFkkHtdsf
d1js8a1 (2682 ) -sp------yGMstlhyTSYDPLFYLHHSNTDRIWSVWQaLQkyrglpyn
d1llaa2 ( 337 ) PdgrfheepgVMsdtSTSLRDpiFYnWHrFIdnIFheYkntlk
d3awua1 ( 199 ) ---------GQMatg-VSPNDPVFWLHHAYVDkLWaeWqrrhp------d
d4z0ya- ( 265 ) pnn------EDMGnfYSAGYDPvFYIHHAnvDRMWklWkellpgHvdit-
aaaaaaaaaaaaaaaaa
360 370 380 390 400
d1hc1a2
d1js8a1 (2725 ) tAnçei------nkLvkpLkPFnldtNpnavTka--hstGatSFdyhkLg
d1llaa2
d3awua1 ( 233 ) saYvptggtpdVVdlnetMkpwn-------------tVrPadLldhta-y
d4z0ya- ( 309 ) ----------dpdWlnASYvFYden-------kdlvrvyn-dCvnldklk
410 420
d1hc1a2
d1js8a1 (2767 ) YdYdnlnFhgmtipeLeehLkeiqh
d1llaa2
d3awua1 ( 269 ) YtFda
d4z0ya- ( 342 ) YnFien
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| Arthropod hemocyanin | Hemocyanin middle domain | Spiny lobster (Panulirus interruptus) [TaxId: 6735] | 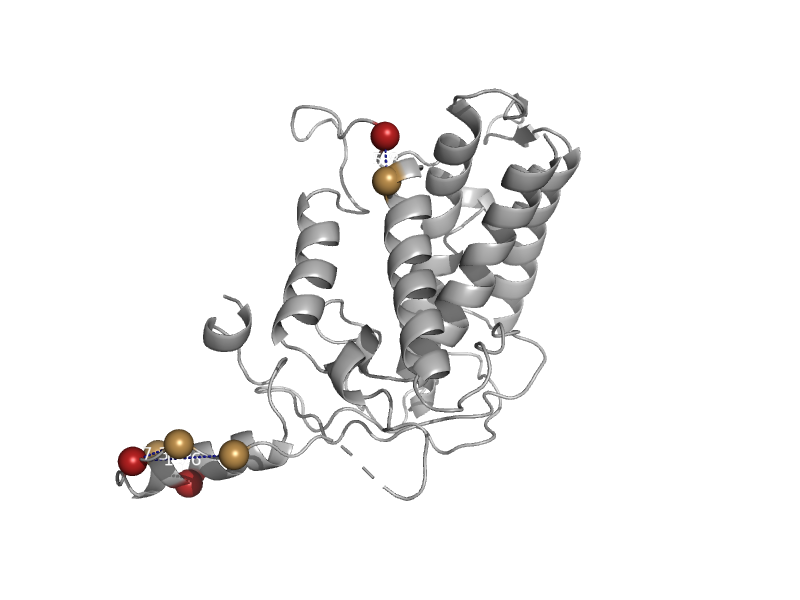 | view | |
| Mollusc hemocyanin | Hemocyanin middle domain | Giant octopus (Octopus dofleini) [TaxId: 267067] | 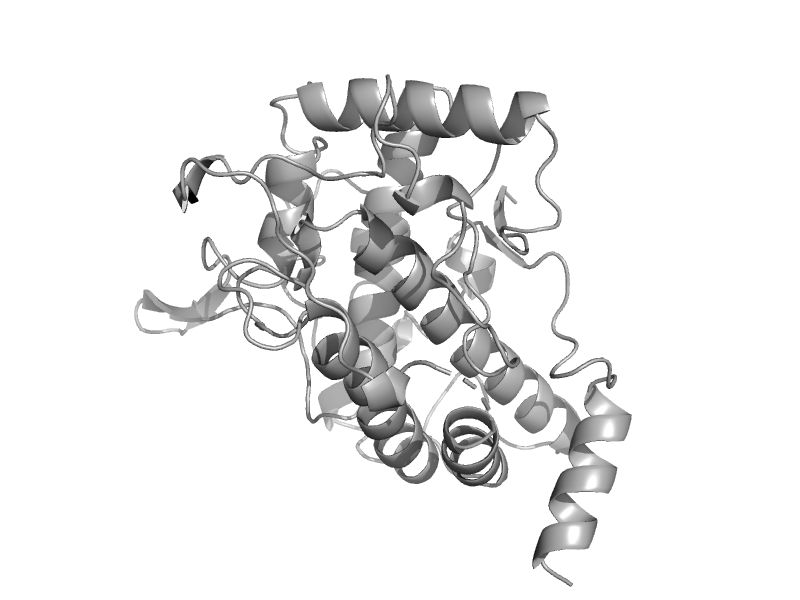 | view | |
| Arthropod hemocyanin | Hemocyanin middle domain | Horseshoe crab (Limulus polyphemus) [TaxId: 6850] | 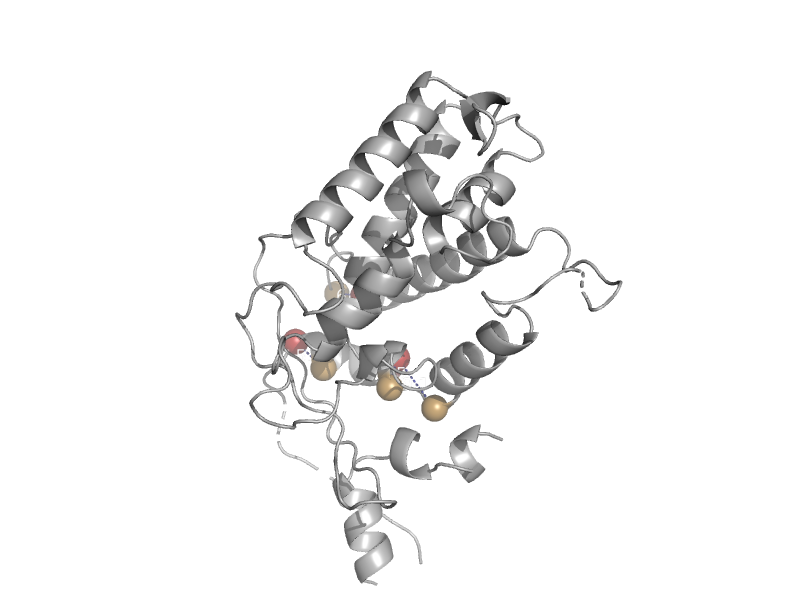 | view | |
| Tyrosinase | Tyrosinase | Streptomyces castaneoglobisporus [TaxId: 79261] | 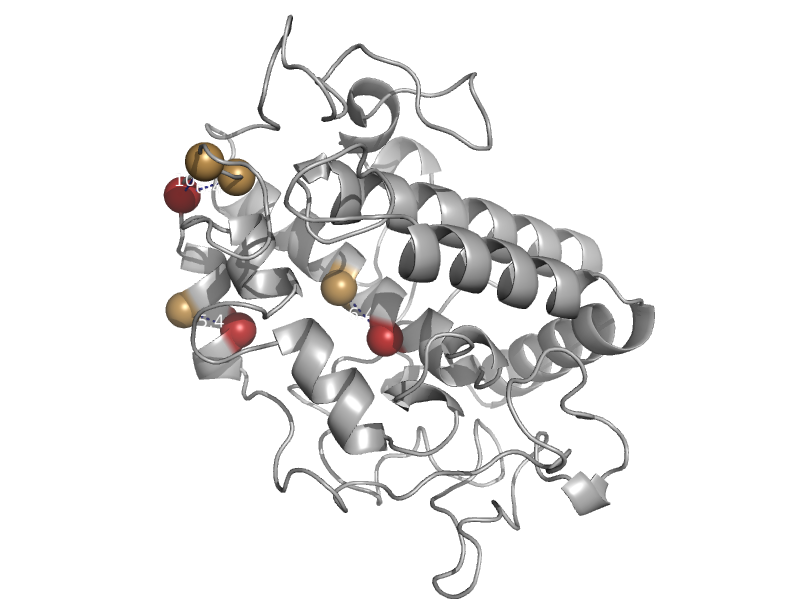 | view | |
| automated matches | automated matches | Coreopsis grandiflora [TaxId: 13449] | 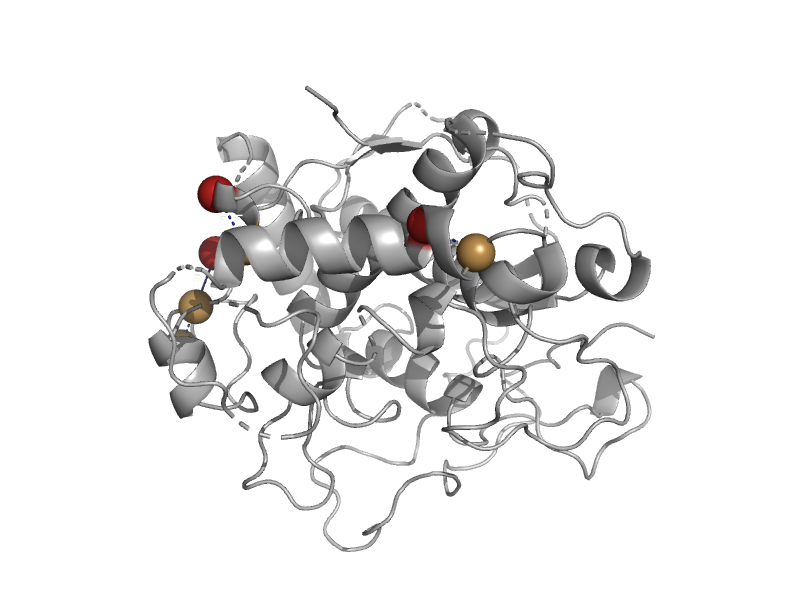 | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | 150 | 160 | 170 | 180 | 190 | 200 | 210 | 220 | 230 | 240 | 250 | 260 | 270 | 280 | 290 | 300 | 310 | 320 | 330 | 340 | 350 | 360 | 370 | 380 | 390 | 400 | 410 | 420 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | P | L | Y | Q | I | T | - | P | H | M | F | T | N | S | E | V | I | D | K | A | Y | S | A | K | M | T | Q | K | P | G | T | F | N | V | S | F | K | N | R | E | Q | R | V | A | Y | F | G | E | D | I | G | M | N | I | H | H | V | T | W | H | M | D | F | P | F | W | W | E | D | S | Y | G | Y | H | L | D | R | K | - | - | G | E | L | F | F | W | V | H | H | Q | L | T | A | R | F | D | F | E | R | L | S | N | W | L | D | - | - | - | P | V | D | E | L | - | H | W | D | - | - | - | - | - | - | R | I | I | R | E | G | F | A | P | L | T | S | Y | K | Y | - | G | G | E | F | P | V | R | P | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | D | N | I | H | F | E | D | V | D | G | V | A | H | V | H | D | L | E | I | T | E | S | R | I | H | E | A | I | D | H | - | - | G | Y | I | T | D | S | D | G | H | T | I | D | I | R | Q | P | K | G | I | E | L | L | G | D | I | I | E | S | S | K | Y | S | S | N | V | Q | Y | Y | G | S | L | H | N | T | A | H | V | M | L | G | R | Q | G | D | P | H | G | K | F | N | L | P | P | G | V | M | E | H | F | E | T | A | T | R | D | P | S | F | F | R | L | H | K | Y | M | D | N | I | F | K | K | H | T | D | S | F | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | A | I | I | R | K | N | V | N | S | L | T | P | S | D | I | K | E | L | R | D | A | M | A | K | V | Q | A | D | - | - | T | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | S | D | N | G | Y | Q | K | I | A | S | Y | H | G | I | P | L | - | S | C | H | Y | - | - | - | E | N | G | T | A | Y | A | C | C | Q | H | G | M | V | T | F | P | N | W | H | R | L | L | T | K | Q | M | E | D | A | L | V | A | K | G | - | - | S | H | V | G | I | P | Y | W | D | W | T | T | T | F | A | N | L | P | V | L | V | T | E | E | K | - | - | - | - | - | - | - | - | - | - | - | - | - | - | D | N | S | F | H | - | - | - | - | H | A | H | I | D | V | A | N | - | - | - | T | D | T | T | R | S | P | R | A | Q | - | L | F | S | F | F | Y | R | Q | I | A | L | A | L | E | Q | - | T | - | - | - | - | - | - | - | D | F | C | - | - | - | - | - | - | - | - | - | - | - | - | - | D | F | E | I | Q | F | E | I | - | - | - | - | - | - | - | - | - | - | - | - | G | H | N | A | I | H | S | W | V | G | G | S | - | - | - | S | P | - | - | - | - | - | - | Y | G | M | S | T | L | H | Y | T | S | Y | D | P | L | F | Y | L | H | H | S | N | T | D | R | I | W | S | V | W | Q | A | L | Q | K | Y | R | G | L | P | Y | N | T | A | N | C | E | I | - | - | - | - | - | - | N | K | L | V | K | P | L | K | P | F | N | L | D | T | N | P | N | A | V | T | K | A | - | - | H | S | T | G | A | T | S | F | D | Y | H | K | L | G | Y | D | Y | D | N | L | N | F | H | G | M | T | I | P | E | L | E | E | H | L | K | E | I | Q | H |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | P | V | Q | E | I | F | - | P | D | K | F | I | P | S | A | A | I | N | E | A | F | K | K | I | - | - | - | - | - | - | L | V | T | - | G | I | L | D | P | E | Y | R | L | A | Y | Y | R | E | D | V | G | I | N | A | H | H | W | H | W | H | L | V | Y | P | S | T | W | N | P | Y | F | G | K | K | K | D | R | K | - | - | G | E | L | F | Y | Y | M | H | Q | Q | M | C | A | R | Y | D | C | E | R | L | S | N | G | M | H | - | - | - | R | M | L | P | F | N | N | F | D | - | - | - | - | - | - | E | P | L | - | A | G | Y | A | P | H | L | T | H | V | A | S | G | K | Y | Y | S | P | R | P | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | D | G | L | K | L | R | D | L | - | G | D | I | E | I | S | E | M | V | R | M | R | E | R | I | L | D | S | I | H | L | - | - | G | Y | V | I | S | E | D | G | S | H | K | T | L | D | E | L | H | G | T | D | I | L | G | A | L | V | E | S | S | Y | E | S | V | N | H | E | Y | Y | G | N | L | H | N | W | G | H | V | T | M | A | R | I | H | D | P | D | G | R | F | H | E | E | P | G | V | M | S | D | T | S | T | S | L | R | D | P | I | F | Y | N | W | H | R | F | I | D | N | I | F | H | E | Y | K | N | T | L | K | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | T | V | R | K | N | Q | A | T | L | T | A | D | E | K | R | R | F | V | A | A | V | L | E | L | K | R | S | - | - | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | R | Y | D | E | F | V | R | T | H | N | E | F | I | M | S | D | T | D | - | - | - | S | - | - | - | - | G | E | R | T | G | H | R | S | P | S | F | L | P | W | H | R | R | F | L | L | D | F | E | Q | A | L | Q | S | V | D | - | - | S | S | V | T | L | P | Y | W | D | W | S | A | D | R | T | V | R | A | S | L | W | - | A | P | D | F | L | G | G | T | G | R | S | T | D | G | R | V | M | D | G | P | F | A | A | S | T | G | N | W | P | I | N | V | R | V | D | S | R | T | Y | L | R | R | S | L | G | G | S | V | A | E | L | P | T | R | A | E | V | E | S | V | L | A | I | - | S | A | Y | D | L | P | P | Y | N | S | A | S | E | - | - | - | - | - | - | - | - | - | - | - | G | F | R | N | H | L | E | G | W | R | G | - | - | - | - | - | - | - | V | N | L | H | N | R | V | H | V | W | V | G | - | - | - | - | - | - | - | - | - | - | - | - | - | G | Q | M | A | T | G | - | V | S | P | N | D | P | V | F | W | L | H | H | A | Y | V | D | K | L | W | A | E | W | Q | R | R | H | P | - | - | - | - | - | - | D | S | A | Y | V | P | T | G | G | T | P | D | V | V | D | L | N | E | T | M | K | P | W | N | - | - | - | - | - | - | - | - | - | - | - | - | - | T | V | R | P | A | D | L | L | D | H | T | A | - | Y | Y | T | F | D | A | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| A | P | I | T | A | P | D | I | T | S | I | C | K | D | A | S | S | G | I | G | N | Q | E | G | A | I | R | T | R | K | C | C | P | P | S | L | G | K | I | K | D | F | F | P | N | D | V | R | M | R | W | P | A | H | G | - | T | K | K | Q | V | D | D | Y | R | R | A | I | A | A | M | R | A | L | P | D | D | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | D | P | R | S | F | V | S | Q | A | K | I | H | C | A | Y | C | N | G | G | Y | T | Q | V | D | S | G | F | P | D | I | D | I | Q | I | H | N | S | W | L | F | F | P | F | H | R | W | Y | L | Y | F | Y | E | R | I | L | G | S | L | I | D | E | P | N | F | A | L | P | Y | W | K | W | D | E | P | K | G | M | P | I | S | I | F | L | G | - | - | - | - | - | - | - | D | A | S | N | P | L | Y | D | Q | Y | R | D | A | N | H | I | E | D | R | V | D | L | D | Y | D | G | D | K | D | - | - | - | I | P | D | Q | Q | V | A | C | N | L | S | T | V | Y | R | D | L | V | R | N | G | V | D | P | T | S | F | F | G | G | K | Y | V | A | G | D | S | P | V | A | N | - | - | - | - | - | G | D | P | S | V | G | S | V | E | A | - | - | - | - | - | - | - | - | - | - | - | - | G | S | T | A | V | H | R | W | V | G | D | P | T | Q | P | N | N | - | - | - | - | - | - | E | D | M | G | N | F | Y | S | A | G | Y | D | P | V | F | Y | I | H | H | A | N | V | D | R | M | W | K | L | W | K | E | L | L | P | G | H | V | D | I | T | - | - | - | - | - | - | - | - | - | - | - | D | P | D | W | L | N | A | S | Y | V | F | Y | D | E | N | - | - | - | - | - | - | - | K | D | L | V | R | V | Y | N | - | D | C | V | N | L | D | K | L | K | Y | N | F | I | E | N | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Member | Chain | UniProtKB | GO term |
|---|---|---|---|
| d1hc1a2 | A | P04254 | GO:0005344 |
| d1hc1a2 | A | P04254 | GO:0005576 |
| d1hc1a2 | A | P04254 | GO:0015671 |
| d1hc1a2 | A | P04254 | GO:0016491 |
| d1hc1a2 | A | P04254 | GO:0046872 |
| d1hc1a2 | A | P04254 | GO:0005344 |
| d1hc1a2 | A | P04254 | GO:0005576 |
| d1hc1a2 | A | P04254 | GO:0015671 |
| d1hc1a2 | A | P04254 | GO:0016491 |
| d1hc1a2 | A | P04254 | GO:0046872 |
| d1js8a1 | A | O61363 | GO:0016491 |
| d1llaa2 | A | P04253 | GO:0005344 |
| d1llaa2 | A | P04253 | GO:0005507 |
| d1llaa2 | A | P04253 | GO:0005576 |
| d1llaa2 | A | P04253 | GO:0015671 |
| d1llaa2 | A | P04253 | GO:0016491 |
| d1llaa2 | A | P04253 | GO:0031404 |
| d1llaa2 | A | P04253 | GO:0046872 |
| d1llaa2 | A | P04253 | GO:0005344 |
| d1llaa2 | A | P04253 | GO:0005507 |
| d1llaa2 | A | P04253 | GO:0005576 |
| d1llaa2 | A | P04253 | GO:0015671 |
| d1llaa2 | A | P04253 | GO:0016491 |
| d1llaa2 | A | P04253 | GO:0031404 |
| d1llaa2 | A | P04253 | GO:0046872 |
