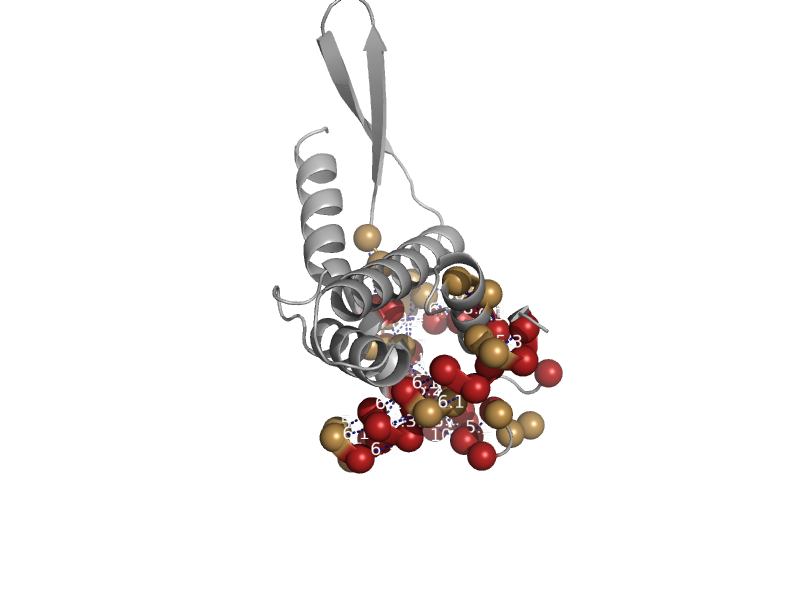
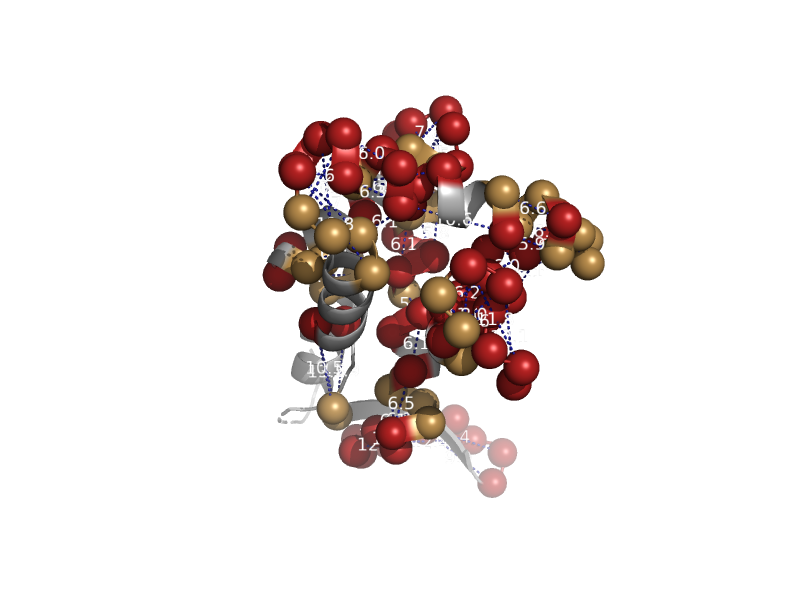
solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1iqva- ( 18 ) ikvMgrWstedVeVkdpsLkpyInleprvhIverLinkVmrsggsskkvr
d1rssa- ( 12 ) lqpdlvygdvlVtaFinkimr---dgkknl
aaaaaaaaa aaa
60 70 80 90 100
d1iqva- ( 103 ) AyevVkeAFkiIekrtgknPIqVLVwAIenAAPredttsvmfggiryhva
d1rssa- ( 39 ) AariFydACkiIqektgqeplkvFkqAVenVkPrmEvrsrrvgganyqvp
aaaaaaaaaaaaaaaa aaaaaaaaaaaa bbbbbbb bbbbbb
110 120 130 140 150
d1iqva- ( 153 ) vdIsplrrLdvALrnIAlgAsakcyrtkmsFaeALAeEIilAAnkdpkSy
d1rssa- ( 89 ) meVsprrqqsLALrWLvqaAnqrp--er-raAvriAhELmdAaegk--gg
b aaaaaaaaaaaaaaaa aaaaaaaaaaaaa a
160 170
d1iqva- ( 203 ) AyskkleieriAessr
d1rssa- ( 134 ) AVkkkedvermAea--hyrw
aaaaaaaaaaaa
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | 150 | 160 | 170 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I | K | V | M | G | R | W | S | T | E | D | V | E | V | K | D | P | S | L | K | P | Y | I | N | L | E | P | R | V | H | I | V | E | R | L | I | N | K | V | M | R | S | G | G | S | S | K | K | V | R | A | Y | E | V | V | K | E | A | F | K | I | I | E | K | R | T | G | K | N | P | I | Q | V | L | V | W | A | I | E | N | A | A | P | R | E | D | T | T | S | V | M | F | G | G | I | R | Y | H | V | A | V | D | I | S | P | L | R | R | L | D | V | A | L | R | N | I | A | L | G | A | S | A | K | C | Y | R | T | K | M | S | F | A | E | A | L | A | E | E | I | I | L | A | A | N | K | D | P | K | S | Y | A | Y | S | K | K | L | E | I | E | R | I | A | E | S | S | R | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | L | Q | P | D | L | V | Y | G | D | V | L | V | T | A | F | I | N | K | I | M | R | - | - | - | D | G | K | K | N | L | A | A | R | I | F | Y | D | A | C | K | I | I | Q | E | K | T | G | Q | E | P | L | K | V | F | K | Q | A | V | E | N | V | K | P | R | M | E | V | R | S | R | R | V | G | G | A | N | Y | Q | V | P | M | E | V | S | P | R | R | Q | Q | S | L | A | L | R | W | L | V | Q | A | A | N | Q | R | P | - | - | E | R | - | R | A | A | V | R | I | A | H | E | L | M | D | A | A | E | G | K | - | - | G | G | A | V | K | K | K | E | D | V | E | R | M | A | E | A | - | - | H | Y | R | W |
| Member | Chain | UniProtKB | GO term |
|---|---|---|---|
| d1iqva_ | A | O59230 | GO:0000028 |
| d1iqva_ | A | O59230 | GO:0003729 |
| d1iqva_ | A | O59230 | GO:0003735 |
| d1iqva_ | A | O59230 | GO:0005840 |
| d1iqva_ | A | O59230 | GO:0006412 |
| d1iqva_ | A | O59230 | GO:0015935 |
| d1iqva_ | A | O59230 | GO:0019843 |
| d1iqva_ | A | O59230 | GO:0022627 |
| d1iqva_ | A | O59230 | GO:1990904 |
| d1rssa_ | A | P17291 | GO:0000028 |
| d1rssa_ | A | P17291 | GO:0000049 |
| d1rssa_ | A | P17291 | GO:0003723 |
| d1rssa_ | A | P17291 | GO:0003729 |
| d1rssa_ | A | P17291 | GO:0003735 |
| d1rssa_ | A | P17291 | GO:0005840 |
| d1rssa_ | A | P17291 | GO:0006412 |
| d1rssa_ | A | P17291 | GO:0015935 |
| d1rssa_ | A | P17291 | GO:0019843 |
| d1rssa_ | A | P17291 | GO:0022627 |
| d1rssa_ | A | P17291 | GO:1990904 |