solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d2g77a1 ( 247 ) elnsiiqriskFdnilkdktiInqqdLrqISwngIpkihrPvVwKLligY
d2g77a2 ( 443 )
60 70 80 90 100
d2g77a1 ( 297 ) LpvntkrqegflqrkrkeYrdslkhTfsdqhsrdiptwhqIeiDIprTnp
d2g77a2 ( 443 )
110 120 130 140 150
d2g77a1 ( 347 ) hipLYqfksVqnSLqrILYLWAirhpasgYvq-----gIndlVTpfFetF
d2g77a2 ( 443 ) gqpgilrqvknLsqlVkridadLynhFqnehVeFiqFAfrWMncl
aaaaaaaaaaaaaaaa aaaaaaaaa
160 170 180 190 200
d2g77a1 ( 392 ) LteyLppsqiddVkikdPstyMvdeqitdLEADTFwCLtklLeqitd---
d2g77a2 ( 488 ) LmrEFqmgTVirMwd-tYlsetssslneFHvfVCAAFLikwsdqLmemdf
333 aaaaaaaaaaaaaaa aa
210 220 230
d2g77a1 ( 439 ) -nyih
d2g77a2 ( 596 ) qeTitfLqnPpTdwtetdIemLLseAfiwQslyka
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| Ypt/Rab-GAP domain of gyp1p, N-terminal domain | Ypt/Rab-GAP domain of gyp1p | Baker's yeast (Saccharomyces cerevisiae) [TaxId: 4932] | 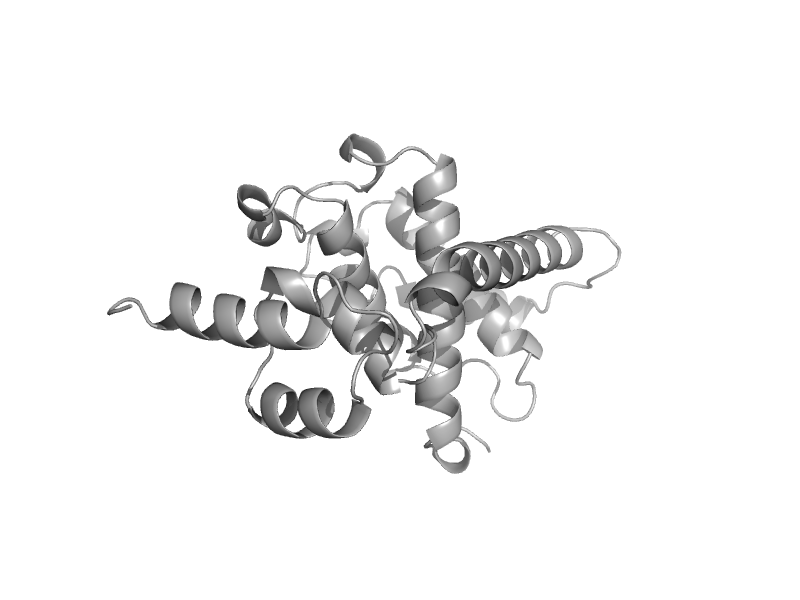 | view | |
| Ypt/Rab-GAP domain of gyp1p, C-terminal domain | Ypt/Rab-GAP domain of gyp1p | Baker's yeast (Saccharomyces cerevisiae) [TaxId: 4932] | 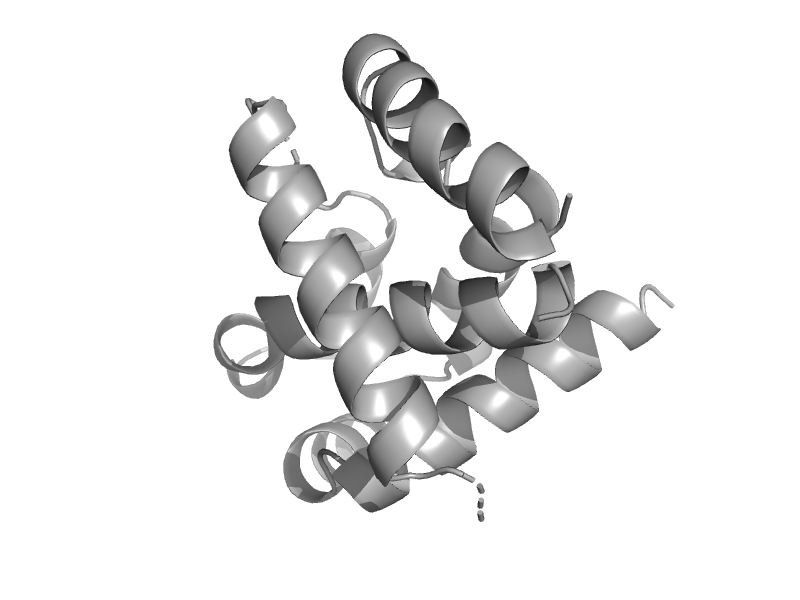 | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | 150 | 160 | 170 | 180 | 190 | 200 | 210 | 220 | 230 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E | L | N | S | I | I | Q | R | I | S | K | F | D | N | I | L | K | D | K | T | I | I | N | Q | Q | D | L | R | Q | I | S | W | N | G | I | P | K | I | H | R | P | V | V | W | K | L | L | I | G | Y | L | P | V | N | T | K | R | Q | E | G | F | L | Q | R | K | R | K | E | Y | R | D | S | L | K | H | T | F | S | D | Q | H | S | R | D | I | P | T | W | H | Q | I | E | I | D | I | P | R | T | N | P | H | I | P | L | Y | Q | F | K | S | V | Q | N | S | L | Q | R | I | L | Y | L | W | A | I | R | H | P | A | S | G | Y | V | Q | - | - | - | - | - | G | I | N | D | L | V | T | P | F | F | E | T | F | L | T | E | Y | L | P | P | S | Q | I | D | D | V | K | I | K | D | P | S | T | Y | M | V | D | E | Q | I | T | D | L | E | A | D | T | F | W | C | L | T | K | L | L | E | Q | I | T | D | - | - | - | - | N | Y | I | H | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | Q | P | G | I | L | R | Q | V | K | N | L | S | Q | L | V | K | R | I | D | A | D | L | Y | N | H | F | Q | N | E | H | V | E | F | I | Q | F | A | F | R | W | M | N | C | L | L | M | R | E | F | Q | M | G | T | V | I | R | M | W | D | - | T | Y | L | S | E | T | S | S | S | L | N | E | F | H | V | F | V | C | A | A | F | L | I | K | W | S | D | Q | L | M | E | M | D | F | Q | E | T | I | T | F | L | Q | N | P | P | T | D | W | T | E | T | D | I | E | M | L | L | S | E | A | F | I | W | Q | S | L | Y | K | A |
| Member | Chain | UniProtKB | GO term |
|---|
