solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1d8ba- ( 11 ) el
d1go3f- ( 1 ) migkkilgeryvTvSEaaimynraqigels--------------------
d1wuda1 ( 530 )
d1y14a- ( 46 ) elialNlSearlvikeaLverrrafkrsqtrekelesIdvlle
d1yt3a2 ( 295 ) lnLmdm
d2cpra1 ( 483 ) kpiftdesylelyrkqkkhln
d2e1fa- (1142 ) qpvis---aqeq
d2hbja1 ( 421 ) keSpwkiLmyqynip
d2ma1a- ( 536 )
60 70 80 90 100
d1d8ba- ( 13 ) nnlrmtYerlrelSlnlGnrmvppvgnFMpdsilkkMAailPmndsaFat
d1go3f- ( 32 ) -------yeqgcaldylqkfakld--ke-eAkLveeLislgidekTAvkI
d1wuda1 ( 530 ) nydrklfakLrklRksiAdesnvppyvVfndatLie-Aeqpita---sel
d1y14a- ( 132 ) qTtggnnkdlkntmqyLTnfSrFRdqet-VgaViqlLkstgLhpfEvaqL
d1yt3a2 ( 301 ) pgyrkAfkaIkslItdvsethkisaelLAsrrQInqLLnwhWkLkpqnnl
d2cpra1 ( 504 ) tqqltAFqllfaWrdktArredesygyvlpnhmMlkiAeelpkepqgiia
d2e1fa- (1151 ) etqivLygkLvearqkhAnkmdvppaiLAtnkiLvdMAkmRPttvenVkr
d2hbja1 ( 436 ) perevLVreLyqwRdliArrddesprfVMpnqLLaALVaytptdvigVvs
d2ma1a- ( 536 ) hdaplfealrawrlqKakelslppytifhdatlktiaelrpgshatlgt
aaaaaaaaaaaaaaaaa aaa aaaaaaaaa
110 120 130 140
d1d8ba- ( 63 ) lg-tve--dkyrrrFkyFkatiadlskkrsse
d1go3f- ( 73 ) AdilPedlddLraiYy---krlpaeeIliVyi
d1wuda1 ( 579 ) svngvg--rklerfgkpfa-lirahvdgd
d1y14a- ( 181 ) GslaCdtAdeAktlIpsLnnkisddeLerILkeLsnletly
d1yt3a2 ( 351 ) PeLIsg--wrgelMaeaLhnlLqeYpq
d2cpra1 ( 554 ) cCnpvp--plvrqqineMhlliqqArempllksevaagvkks
d2e1fa- (1201 ) Id-gVs--egkAamLapLLevIkhfCqtn----svqtdlfss
d2hbja1 ( 486 ) LtngVt--ehVrqnAklLAnlIrdalrnikntn
d2ma1a- ( 585 ) vsgvgg--rklaaYgdevlqvvrdssg--------------g
aaaaa aa aaa
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| HRDC domain from RecQ helicase | HRDC domain from helicases | Baker's yeast (Saccharomyces cerevisiae) [TaxId: 4932] | 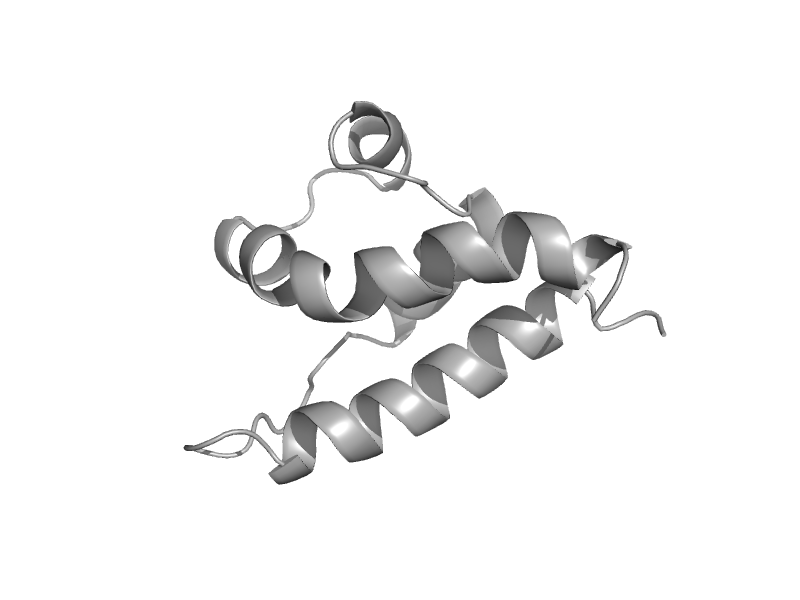 | view | |
| RNA polymerase II subunit RBP4 (RpoF) | RNA polymerase II subunit RBP4 (RpoF) | Methanococcus jannaschii [TaxId: 2190] | 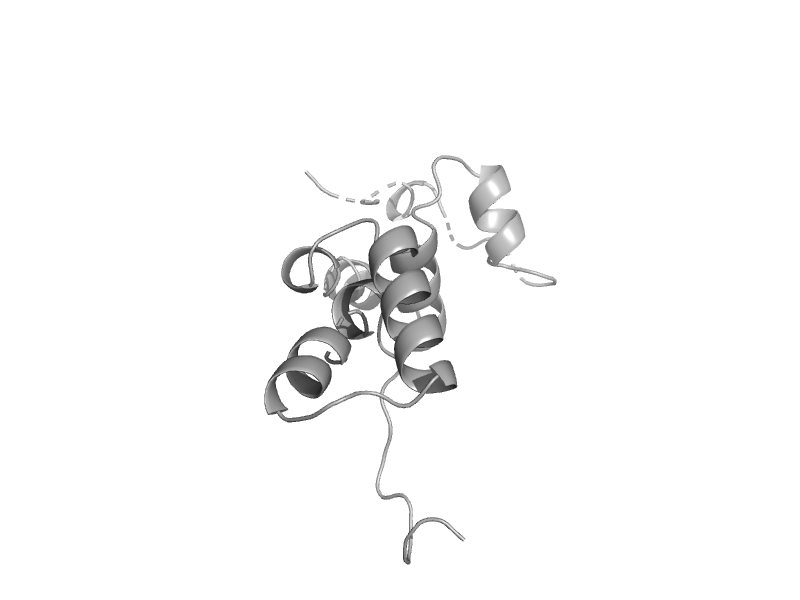 | view | |
| HRDC domain from RecQ helicase | HRDC domain from helicases | Escherichia coli [TaxId: 562] | 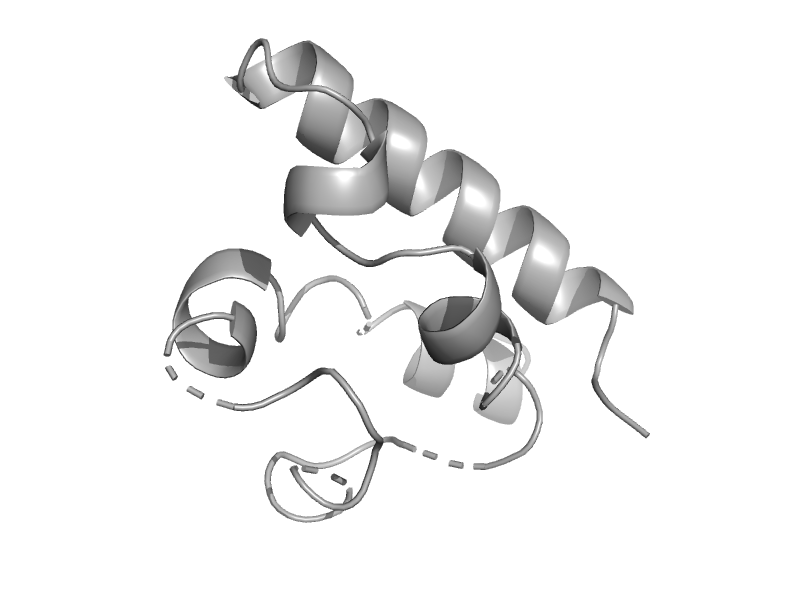 | view | |
| RNA polymerase II subunit RBP4 (RpoF) | RNA polymerase II subunit RBP4 (RpoF) | Baker's yeast (Saccharomyces cerevisiae) [TaxId: 4932] | 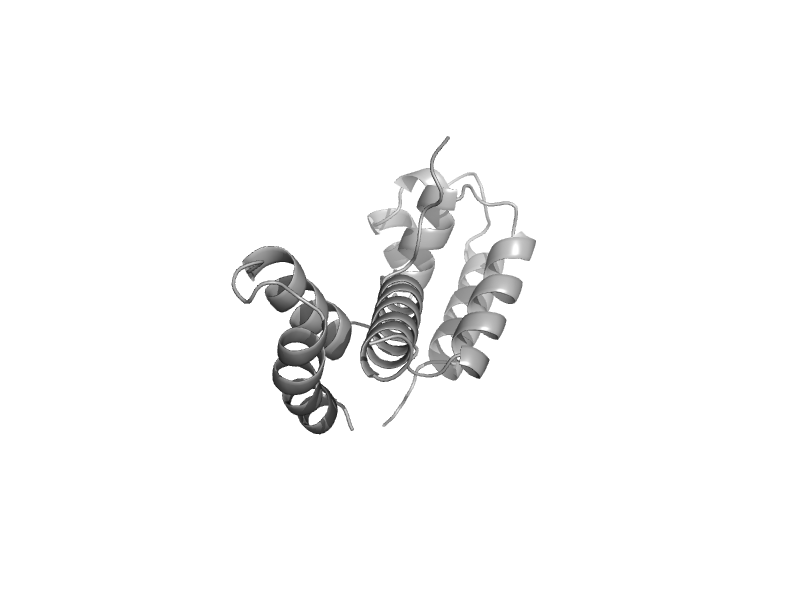 | view | |
| Ribonuclease D | RNase D C-terminal domains | Escherichia coli [TaxId: 562] | 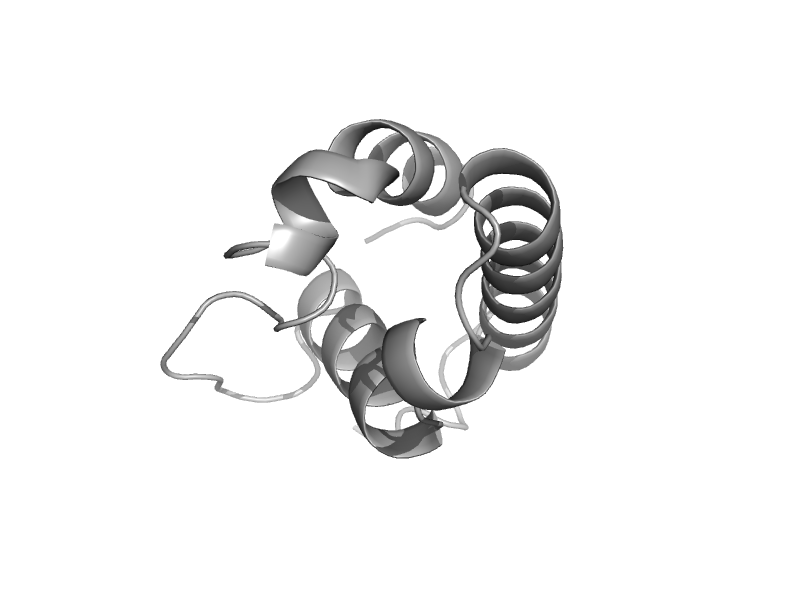 | view | |
| Exosome component 10, EXOSC10 | EXOSC10 HRDC domain-like | Human (Homo sapiens) [TaxId: 9606] | 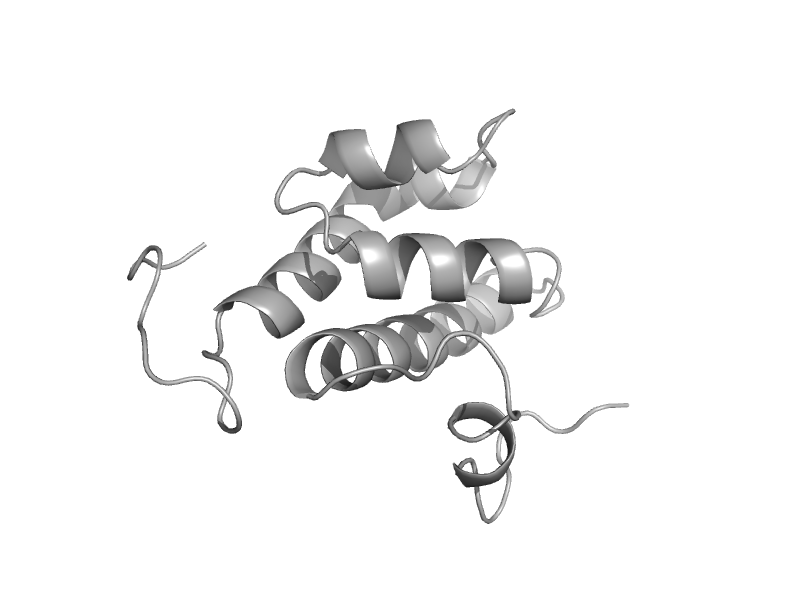 | view | |
| automated matches | HRDC domain from helicases | Human (Homo sapiens) [TaxId: 9606] |  | view | |
| Exosome complex exonuclease RRP6 domain | EXOSC10 HRDC domain-like | Baker's yeast (Saccharomyces cerevisiae) [TaxId: 4932] | 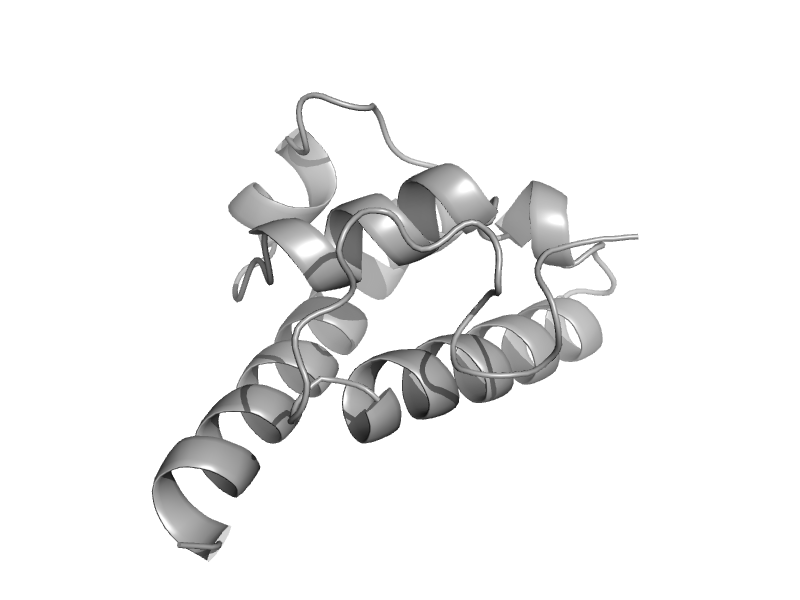 | view | |
| automated matches | automated matches | Deinococcus radiodurans [TaxId: 243230] | 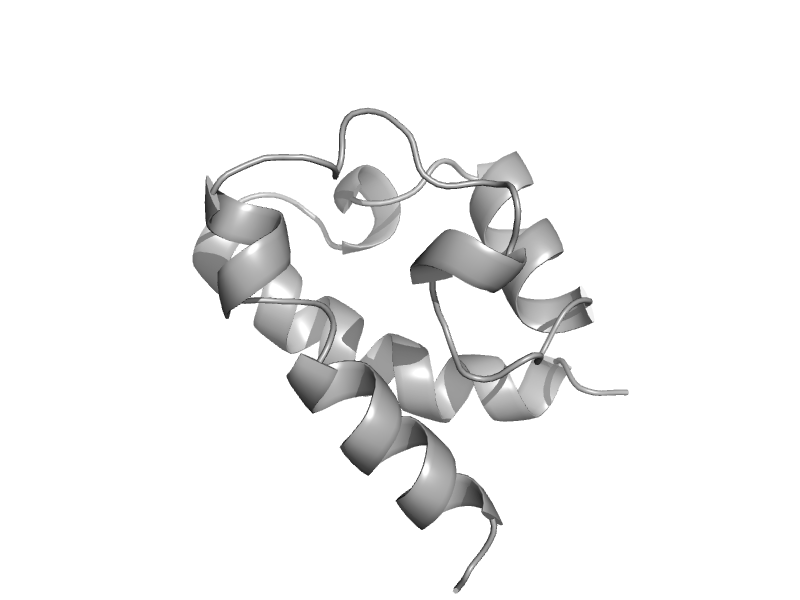 | view |
| Domain ID | Name |
|---|---|
| d7f4gd- | 7f4g D: |
| d1yt3a1 | 1yt3 A:194-294 |
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | E | L | N | N | L | R | M | T | Y | E | R | L | R | E | L | S | L | N | L | G | N | R | M | V | P | P | V | G | N | F | M | P | D | S | I | L | K | K | M | A | A | I | L | P | M | N | D | S | A | F | A | T | L | G | - | T | V | E | - | - | D | K | Y | R | R | R | F | K | Y | F | K | A | T | I | A | D | L | S | K | K | R | S | S | E | - | - | - | - | - | - | - | - | - | - |
| M | I | G | K | K | I | L | G | E | R | Y | V | T | V | S | E | A | A | I | M | Y | N | R | A | Q | I | G | E | L | S | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Y | E | Q | G | C | A | L | D | Y | L | Q | K | F | A | K | L | D | - | - | K | E | - | E | A | K | L | V | E | E | L | I | S | L | G | I | D | E | K | T | A | V | K | I | A | D | I | L | P | E | D | L | D | D | L | R | A | I | Y | Y | - | - | - | K | R | L | P | A | E | E | I | L | I | V | Y | I | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | N | Y | D | R | K | L | F | A | K | L | R | K | L | R | K | S | I | A | D | E | S | N | V | P | P | Y | V | V | F | N | D | A | T | L | I | E | - | A | E | Q | P | I | T | A | - | - | - | S | E | L | S | V | N | G | V | G | - | - | R | K | L | E | R | F | G | K | P | F | A | - | L | I | R | A | H | V | D | G | D | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | E | L | I | A | L | N | L | S | E | A | R | L | V | I | K | E | A | L | V | E | R | R | R | A | F | K | R | S | Q | T | R | E | K | E | L | E | S | I | D | V | L | L | E | Q | T | T | G | G | N | N | K | D | L | K | N | T | M | Q | Y | L | T | N | F | S | R | F | R | D | Q | E | T | - | V | G | A | V | I | Q | L | L | K | S | T | G | L | H | P | F | E | V | A | Q | L | G | S | L | A | C | D | T | A | D | E | A | K | T | L | I | P | S | L | N | N | K | I | S | D | D | E | L | E | R | I | L | K | E | L | S | N | L | E | T | L | Y | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | L | N | L | M | D | M | P | G | Y | R | K | A | F | K | A | I | K | S | L | I | T | D | V | S | E | T | H | K | I | S | A | E | L | L | A | S | R | R | Q | I | N | Q | L | L | N | W | H | W | K | L | K | P | Q | N | N | L | P | E | L | I | S | G | - | - | W | R | G | E | L | M | A | E | A | L | H | N | L | L | Q | E | Y | P | Q | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | K | P | I | F | T | D | E | S | Y | L | E | L | Y | R | K | Q | K | K | H | L | N | T | Q | Q | L | T | A | F | Q | L | L | F | A | W | R | D | K | T | A | R | R | E | D | E | S | Y | G | Y | V | L | P | N | H | M | M | L | K | I | A | E | E | L | P | K | E | P | Q | G | I | I | A | C | C | N | P | V | P | - | - | P | L | V | R | Q | Q | I | N | E | M | H | L | L | I | Q | Q | A | R | E | M | P | L | L | K | S | E | V | A | A | G | V | K | K | S |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Q | P | V | I | S | - | - | - | A | Q | E | Q | E | T | Q | I | V | L | Y | G | K | L | V | E | A | R | Q | K | H | A | N | K | M | D | V | P | P | A | I | L | A | T | N | K | I | L | V | D | M | A | K | M | R | P | T | T | V | E | N | V | K | R | I | D | - | G | V | S | - | - | E | G | K | A | A | M | L | A | P | L | L | E | V | I | K | H | F | C | Q | T | N | - | - | - | - | S | V | Q | T | D | L | F | S | S |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | K | E | S | P | W | K | I | L | M | Y | Q | Y | N | I | P | P | E | R | E | V | L | V | R | E | L | Y | Q | W | R | D | L | I | A | R | R | D | D | E | S | P | R | F | V | M | P | N | Q | L | L | A | A | L | V | A | Y | T | P | T | D | V | I | G | V | V | S | L | T | N | G | V | T | - | - | E | H | V | R | Q | N | A | K | L | L | A | N | L | I | R | D | A | L | R | N | I | K | N | T | N | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | H | D | A | P | L | F | E | A | L | R | A | W | R | L | Q | K | A | K | E | L | S | L | P | P | Y | T | I | F | H | D | A | T | L | K | T | I | A | E | L | R | P | G | S | H | A | T | L | G | T | V | S | G | V | G | G | - | - | R | K | L | A | A | Y | G | D | E | V | L | Q | V | V | R | D | S | S | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G |
