solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1c02a- ( 2 ) stIpseiInwtiLneiismdd----------------
d1i5na- ( 5 )
d1sr2a- ( 775 ) mqeavlqlievqla
d1tqga1 ( 4 )
d1wn0a1 ( 9 ) qlnallssMfasglVd-eqFqqLQmlqedg--------------
d1y6da- ( 1 )
d2a0ba- ( 658 ) ksealLdipmLeqylel------------------
d2ooca1 ( 8 )
d4g78a1 ( 1 ) mdhlhrklrdheaaMfqqgyLd-dqFsqlqkl--qd--------------
60 70 80 90 100
d1c02a- ( 23 ) ----------dds-dfSk--glIiqFi-----dqAqtTFaqMqrQLdg--
d1i5na- ( 5 ) isdfyqtFf-----deAdellad-eqhLldL-
d1sr2a- ( 789 ) qeevtesplggdenaqlhasgyyalFv-----dtvpdDvkrlyteaa---
d1tqga1 ( 4 ) eylgvFv-----detkeylqnlndtllele
d1wn0a1 ( 38 ) ----------gtp-gfVa--evVtlFc-----ddAdrIIseLaalLdqp-
d1y6da- ( 1 ) mntdvlnqqkieelSaeiGsdnvpvlldiflgemdsyi
d2a0ba- ( 675 ) -----------vgpklIt--dgLavFe-----kmMpgyVsvLesnlt---
d2ooca1 ( 8 ) gaVdfayLegfAa-gdfavVdevLalFreqAalwa
d4g78a1 ( 34 ) ----------dtspdfVi--evMtmFf-----ddSekllnnMsraLeqv-
aaaaaa aaaaaaaaaaaaaaa
110 120 130 140 150
d1c02a- ( 53 ) -------eknlteLdnlGhfL-kgsSaalGLqRIa-wVCerIQnLGrkmq
d1i5na- ( 31 ) -vpespdaeqLnaifraahsI-kggAgtfgftilq-eTth-lenLldear
d1sr2a- ( 831 ) -------tsdfaalaqtAhrl-kgvFamlnlvpGk-qlCetlehlir---
d1tqga1 ( 29 ) knpe--dmelineAfralhtl-kgmAgtmgfssMa-klChtlEnildkAr
d1wn0a1 ( 69 ) -------iVdfdkVdayVhqL-kgsSasVgAqkVk-ftCmqFrqlCq---
d1y6da- ( 39 ) gtltelqgSeqllylkEishAlksSAasFGadrlceraiaidkkakAnql
d2a0ba- ( 704 ) -------aqdkkgIveEGhkI-kgaAgsvGLrhLq-qLGqqIqsp---dl
d2ooca1 ( 42 ) pLdpthp-----gwkdAVhtV-kgaArgvGAfnLG--vCrCeag------
d4g78a1 ( 66 ) -------pvnfkqIdahAhqQ-kgsSasVGAarVk-nvCgtFrnfCe---
aaaaaaaaaaa aaaaaa aaaa aaaa a aaa
160 170 180 190 200
d1c02a- ( 94 ) hfFpnkteLvntLsdksiinginideddeeikIqvddkdenSiyLilIak
d1i5na- ( 78 ) rge-----------------------------------qLntdiinlfle
d1sr2a- ( 869 ) -----------------------------------------ekdvpgiek
d1tqga1 ( 75 ) nsei----------------------------------kitsdlldkifa
d1wn0a1 ( 107 ) -----------------------------------------dknrdgCim
d1y6da- ( 89 ) qe--------------------------------------qgmetsEMla
d2a0ba- ( 742 ) p-----------------------------------------aWednVge
d2ooca1 ( 81 ) -----------------------------------------qeslegVrt
d4g78a1 ( 104 ) -----------------------------------------aqnleGCvr
aaaaaa
210 220 230
d1c02a- ( 144 ) ALnQSrLEfkLAri-----eLskyyntnl
d1i5na- ( 94 ) tkd-iqeQLdayknseePdaasfeyICnaLrqlalea
d1sr2a- ( 878 ) yisdidsyvksll
d1tqga1 ( 91 ) Gvdmitrmvdkivs
d1wn0a1 ( 116 ) aLavVrneFydLrn-----kFqtMlqleq
d1y6da- ( 101 ) llhitrdaYrswtn
d2a0ba- ( 751 ) wIeeMkeeWrhDVe-----vLkawVakat
d2ooca1 ( 90 ) aLdaAlldIaayah-----eqalslk
d4g78a1 ( 113 ) clqqLqqeyslLkn-----nLkyLfkLqqeiktagrs
aaaaaaaaaaaa
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| Phosphorelay protein ypd1 | Phosphorelay protein-like | Baker's yeast (Saccharomyces cerevisiae) [TaxId: 4932] | 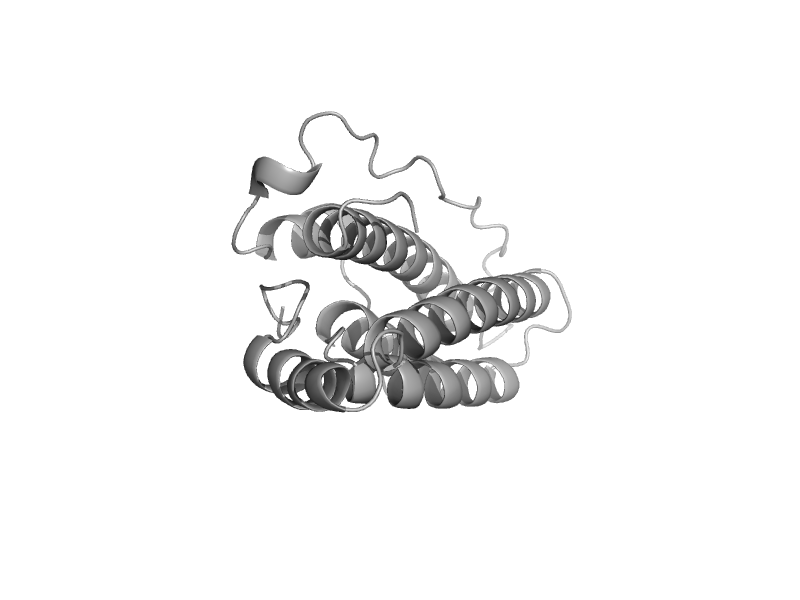 | view | |
| Chemotaxis protein CheA P1 domain | Chemotaxis protein CheA P1 domain | Salmonella typhimurium [TaxId: 90371] | 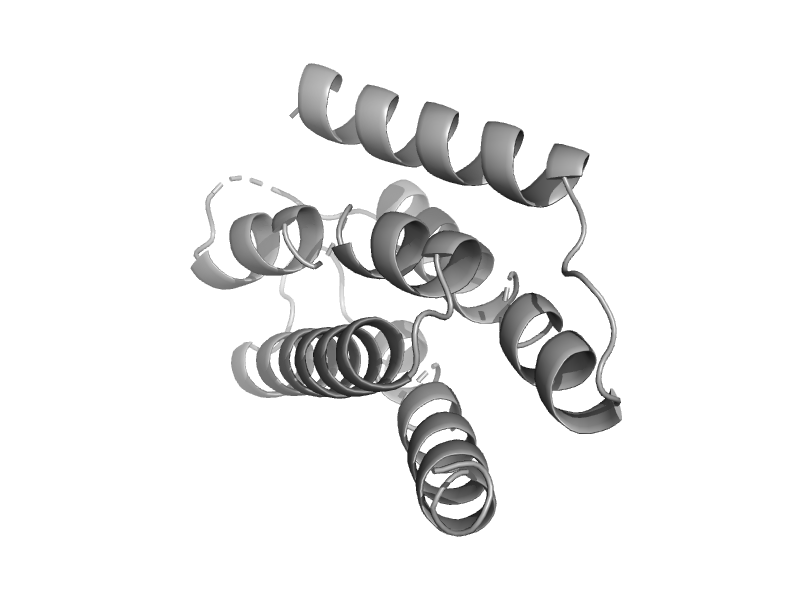 | view | |
| Sensor-like histidine kinase YojN, C-terminal domain | Sensor-like histidine kinase YojN, C-terminal domain | Escherichia coli [TaxId: 562] | 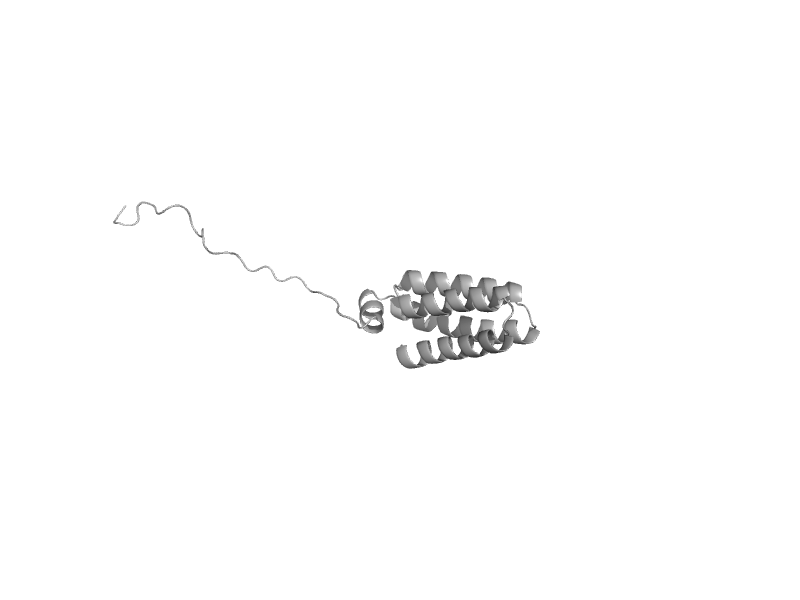 | view | |
| Chemotaxis protein CheA P1 domain | Chemotaxis protein CheA P1 domain | Thermotoga maritima [TaxId: 2336] | 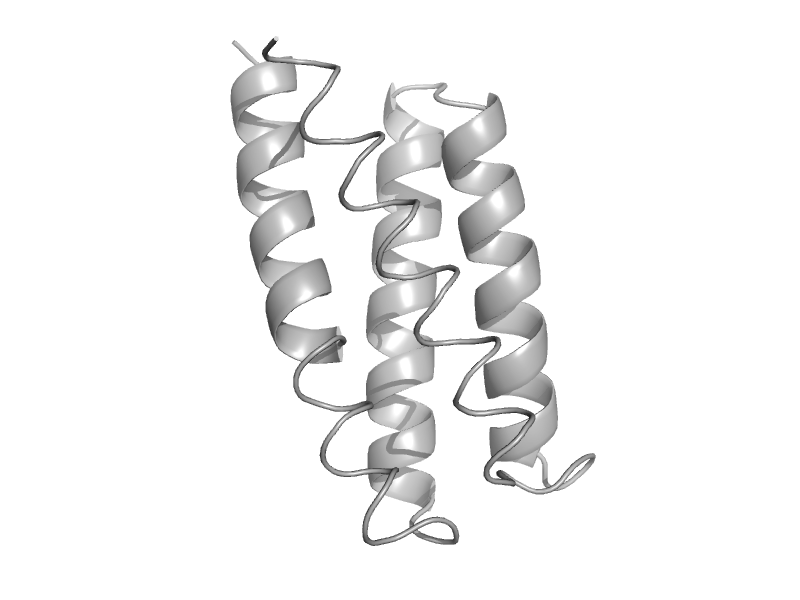 | view | |
| Histidine-containing phosphotransfer protein HP2 | Phosphorelay protein-like | Maize (Zea mays) [TaxId: 4577] | 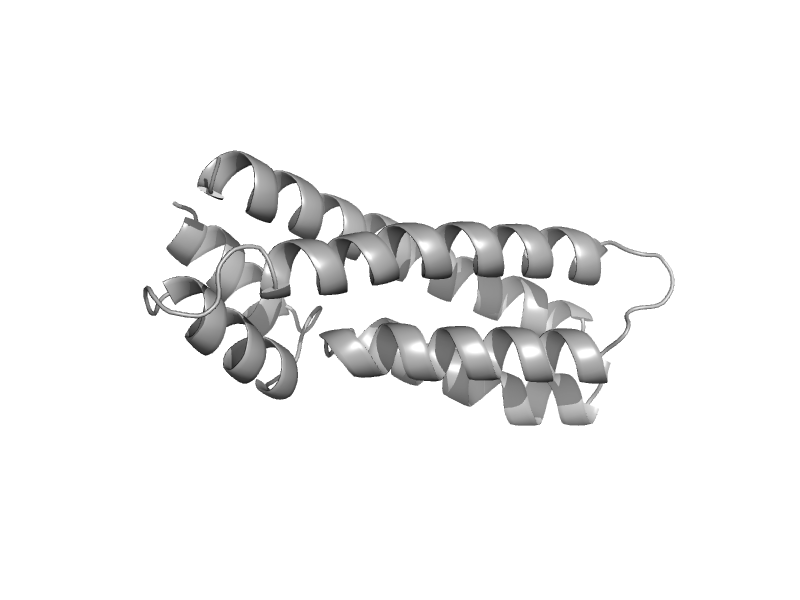 | view | |
| Phosphorelay protein luxU | Phosphorelay protein luxU | Vibrio harveyi [TaxId: 669] | 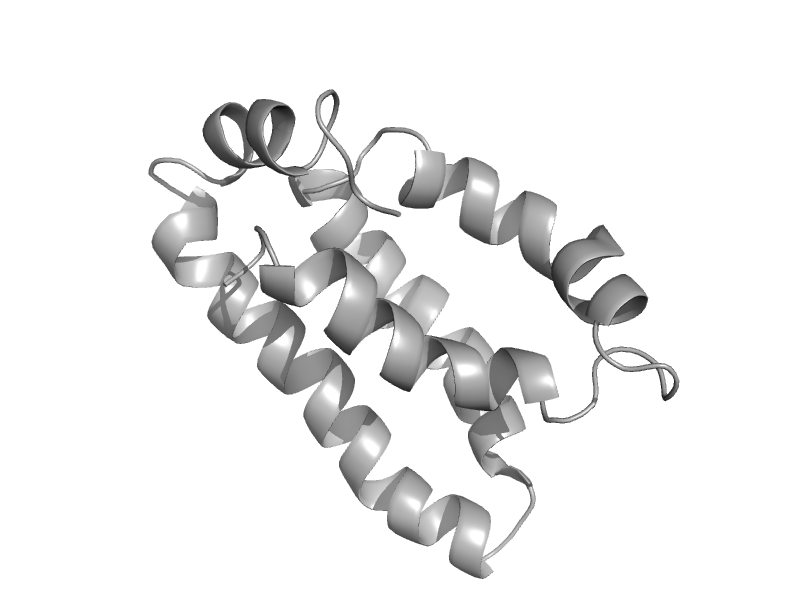 | view | |
| Aerobic respiration control sensor protein, ArcB | Aerobic respiration control sensor protein, ArcB | Escherichia coli [TaxId: 562] | 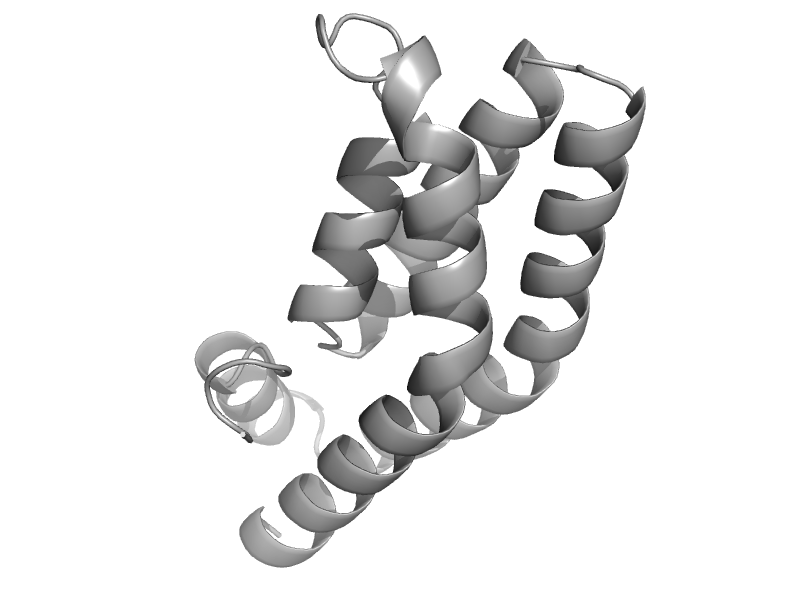 | view | |
| Histidine phosphotransferase ShpA | SphA-like | Caulobacter crescentus [TaxId: 155892] | 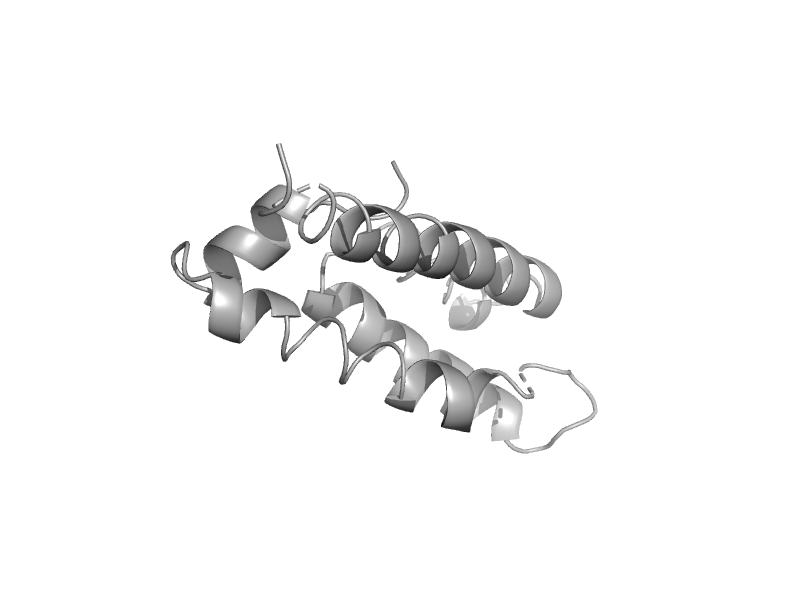 | view | |
| automated matches | automated matches | Medicago truncatula [TaxId: 3880] |  | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | 150 | 160 | 170 | 180 | 190 | 200 | 210 | 220 | 230 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | - | - | - | - | - | - | - | - | - | - | - | - | S | T | I | P | S | E | I | I | N | W | T | I | L | N | E | I | I | S | M | D | D | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | D | D | S | - | D | F | S | K | - | - | G | L | I | I | Q | F | I | - | - | - | - | - | D | Q | A | Q | T | T | F | A | Q | M | Q | R | Q | L | D | G | - | - | - | - | - | - | - | - | - | E | K | N | L | T | E | L | D | N | L | G | H | F | L | - | K | G | S | S | A | A | L | G | L | Q | R | I | A | - | W | V | C | E | R | I | Q | N | L | G | R | K | M | Q | H | F | F | P | N | K | T | E | L | V | N | T | L | S | D | K | S | I | I | N | G | I | N | I | D | E | D | D | E | E | I | K | I | Q | V | D | D | K | D | E | N | S | I | Y | L | I | L | I | A | K | A | L | N | Q | S | R | L | E | F | K | L | A | R | I | - | - | - | - | - | E | L | S | K | Y | Y | N | T | N | L | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | I | S | D | F | Y | Q | T | F | F | - | - | - | - | - | D | E | A | D | E | L | L | A | D | - | E | Q | H | L | L | D | L | - | - | V | P | E | S | P | D | A | E | Q | L | N | A | I | F | R | A | A | H | S | I | - | K | G | G | A | G | T | F | G | F | T | I | L | Q | - | E | T | T | H | - | L | E | N | L | L | D | E | A | R | R | G | E | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Q | L | N | T | D | I | I | N | L | F | L | E | T | K | D | - | I | Q | E | Q | L | D | A | Y | K | N | S | E | E | P | D | A | A | S | F | E | Y | I | C | N | A | L | R | Q | L | A | L | E | A |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | M | Q | E | A | V | L | Q | L | I | E | V | Q | L | A | Q | E | E | V | T | E | S | P | L | G | G | D | E | N | A | Q | L | H | A | S | G | Y | Y | A | L | F | V | - | - | - | - | - | D | T | V | P | D | D | V | K | R | L | Y | T | E | A | A | - | - | - | - | - | - | - | - | - | - | T | S | D | F | A | A | L | A | Q | T | A | H | R | L | - | K | G | V | F | A | M | L | N | L | V | P | G | K | - | Q | L | C | E | T | L | E | H | L | I | R | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | E | K | D | V | P | G | I | E | K | Y | I | S | D | I | D | S | Y | V | K | S | L | L | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | E | Y | L | G | V | F | V | - | - | - | - | - | D | E | T | K | E | Y | L | Q | N | L | N | D | T | L | L | E | L | E | K | N | P | E | - | - | D | M | E | L | I | N | E | A | F | R | A | L | H | T | L | - | K | G | M | A | G | T | M | G | F | S | S | M | A | - | K | L | C | H | T | L | E | N | I | L | D | K | A | R | N | S | E | I | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | K | I | T | S | D | L | L | D | K | I | F | A | G | V | D | M | I | T | R | M | V | D | K | I | V | S | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | Q | L | N | A | L | L | S | S | M | F | A | S | G | L | V | D | - | E | Q | F | Q | Q | L | Q | M | L | Q | E | D | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | T | P | - | G | F | V | A | - | - | E | V | V | T | L | F | C | - | - | - | - | - | D | D | A | D | R | I | I | S | E | L | A | A | L | L | D | Q | P | - | - | - | - | - | - | - | - | I | V | D | F | D | K | V | D | A | Y | V | H | Q | L | - | K | G | S | S | A | S | V | G | A | Q | K | V | K | - | F | T | C | M | Q | F | R | Q | L | C | Q | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | D | K | N | R | D | G | C | I | M | A | L | A | V | V | R | N | E | F | Y | D | L | R | N | - | - | - | - | - | K | F | Q | T | M | L | Q | L | E | Q | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | M | N | T | D | V | L | N | Q | Q | K | I | E | E | L | S | A | E | I | G | S | D | N | V | P | V | L | L | D | I | F | L | G | E | M | D | S | Y | I | G | T | L | T | E | L | Q | G | S | E | Q | L | L | Y | L | K | E | I | S | H | A | L | K | S | S | A | A | S | F | G | A | D | R | L | C | E | R | A | I | A | I | D | K | K | A | K | A | N | Q | L | Q | E | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Q | G | M | E | T | S | E | M | L | A | L | L | H | I | T | R | D | A | Y | R | S | W | T | N | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | K | S | E | A | L | L | D | I | P | M | L | E | Q | Y | L | E | L | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | V | G | P | K | L | I | T | - | - | D | G | L | A | V | F | E | - | - | - | - | - | K | M | M | P | G | Y | V | S | V | L | E | S | N | L | T | - | - | - | - | - | - | - | - | - | - | A | Q | D | K | K | G | I | V | E | E | G | H | K | I | - | K | G | A | A | G | S | V | G | L | R | H | L | Q | - | Q | L | G | Q | Q | I | Q | S | P | - | - | - | D | L | P | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | A | W | E | D | N | V | G | E | W | I | E | E | M | K | E | E | W | R | H | D | V | E | - | - | - | - | - | V | L | K | A | W | V | A | K | A | T | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | A | V | D | F | A | Y | L | E | G | F | A | A | - | G | D | F | A | V | V | D | E | V | L | A | L | F | R | E | Q | A | A | L | W | A | P | L | D | P | T | H | P | - | - | - | - | - | G | W | K | D | A | V | H | T | V | - | K | G | A | A | R | G | V | G | A | F | N | L | G | - | - | V | C | R | C | E | A | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Q | E | S | L | E | G | V | R | T | A | L | D | A | A | L | L | D | I | A | A | Y | A | H | - | - | - | - | - | E | Q | A | L | S | L | K | - | - | - | - | - | - | - | - | - | - | - |
| M | D | H | L | H | R | K | L | R | D | H | E | A | A | M | F | Q | Q | G | Y | L | D | - | D | Q | F | S | Q | L | Q | K | L | - | - | Q | D | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | D | T | S | P | D | F | V | I | - | - | E | V | M | T | M | F | F | - | - | - | - | - | D | D | S | E | K | L | L | N | N | M | S | R | A | L | E | Q | V | - | - | - | - | - | - | - | - | P | V | N | F | K | Q | I | D | A | H | A | H | Q | Q | - | K | G | S | S | A | S | V | G | A | A | R | V | K | - | N | V | C | G | T | F | R | N | F | C | E | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | A | Q | N | L | E | G | C | V | R | C | L | Q | Q | L | Q | Q | E | Y | S | L | L | K | N | - | - | - | - | - | N | L | K | Y | L | F | K | L | Q | Q | E | I | K | T | A | G | R | S |
| Member | Chain | UniProtKB | GO term |
|---|---|---|---|
| d1c02a_ | A | Q07688 | GO:0000160 |
| d1c02a_ | A | Q07688 | GO:0005515 |
| d1c02a_ | A | Q07688 | GO:0005634 |
| d1c02a_ | A | Q07688 | GO:0005737 |
| d1c02a_ | A | Q07688 | GO:0007234 |
| d1c02a_ | A | Q07688 | GO:0009927 |
| d1c02a_ | A | Q07688 | GO:0016772 |
| d1c02a_ | A | Q07688 | GO:0043424 |
| d1i5na_ | A | P09384 | GO:0000160 |
| d1sr2a_ | A | P39838 | GO:0000160 |
| d1wn0a1 | A | Q9SLX1 | GO:0000160 |
| d1wn0a1 | A | Q9SLX1 | GO:0005515 |
| d1wn0a1 | A | Q9SLX1 | GO:0005634 |
| d1wn0a1 | A | Q9SLX1 | GO:0005737 |
| d1wn0a1 | A | Q9SLX1 | GO:0005829 |
| d1wn0a1 | A | Q9SLX1 | GO:0009736 |
| d1wn0a1 | A | Q9SLX1 | GO:0009927 |
| d1wn0a1 | A | Q9SLX1 | GO:0043424 |
| d1wn0a1 | A | Q9SLX1 | GO:0080038 |
| d1y6da_ | A | P0C5S4 | GO:0000160 |
| d2a0ba_ | A | P0AEC3 | GO:0000160 |
| d2ooca1 | A | Q9A980 | GO:0000160 |
| d2ooca1 | A | Q9A980 | GO:0016740 |
