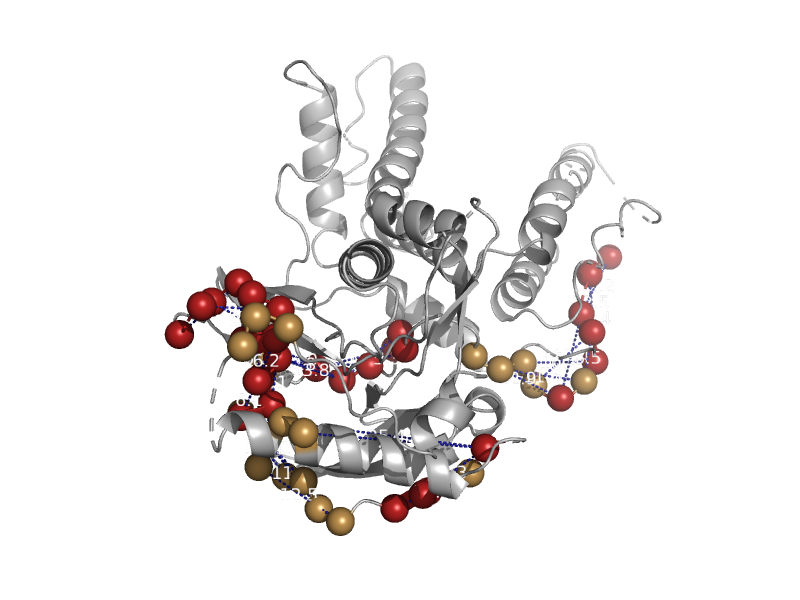
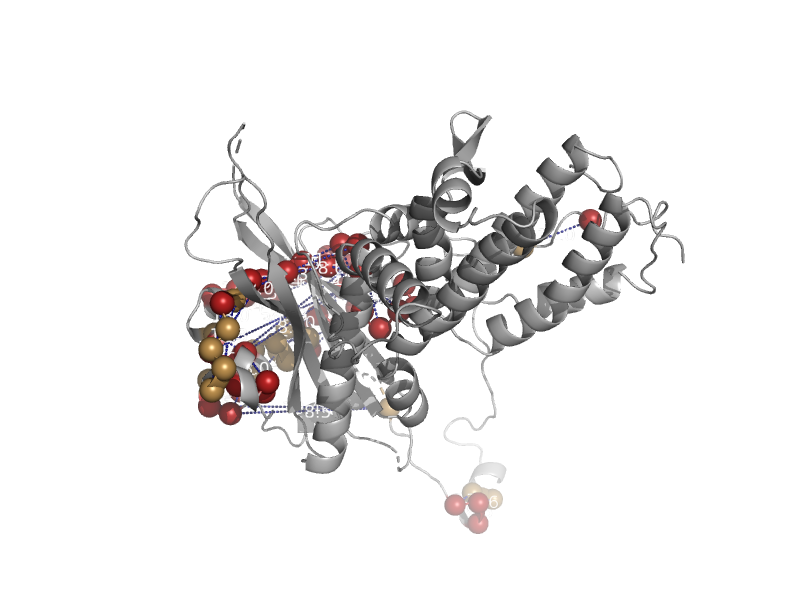
solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d4iuja- ( 266 ) rpLlpdgppCSQrSkFLLmdalkLsiedPshgegipLyDA
d5d9aa3 ( 238 ) ikHyeqnkfrqIrlpkgpmApYtHkFLMeEAwmftkisdperSrageiLI
bbbb aaa
60 70 80 90 100
d4iuja- ( 308 ) ICmrt--ffgwkePnvvkphkGinpnYllSWkqVlaElqdieniprtknM
d5d9aa3 ( 288 ) dffkkgnlsairPkdkplqGk-ypihyknLWnqIkaAIad------rtmV
aaaaaaaaaaaaaaaaa
110 120 130 140 150
d4iuja- ( 361 ) kktsqLkwALgee-----------------------------lrslasWI
d5d9aa3 ( 331 ) inendhseflggraskkipeisltqdvitteglkqsenklpeprsfPrwF
aaaaa aa
160 170 180 190 200
d4iuja- ( 408 ) qnEFnkACeltds-sWIe-----ldeIgedaap-----ihiasmRrnyFT
d5d9aa3 ( 383 ) naEwmwAikdsdltgWVpmaeyp---------padneledyaehlnktMe
aaaaaaaa aaaaaaaaaa
210 220 230 240 250
d4iuja- ( 448 ) aeVshCrATEYImKGvyIntaLlnAScaamddFqLIPMISkCrte-----
d5d9aa3 ( 424 ) gvLqgTNCArEMGKCilTvgaLmtECrlfpgkiKVVPIyArSkerksmqe
aaa aaaaaaaaaaaaaaaaaaaaaa bbbbbbbbb
260 270 280 290 300
d4iuja- ( 494 ) ----grrkTNLYGFIIKGrShLndtdvVnFVSMEFSltdPrlephKWekY
d5d9aa3 ( 474 ) glpvpsemDCLFGICVKSksh---dgmytIITFEFSireP--nlekHqkY
bbbbbbbbbbb bbbbbbbbb
310 320 330 340 350
d4iuja- ( 541 ) CVLeVGdmllrsaighvsrpMFLYVrtngtskikMkwgmeMrrcLlqSLq
d5d9aa3 ( 523 ) TVFeAghttvrm--kkrevpLyLYCRttaLskikNdwlskArrcfitTmd
bbbbbbbbb bbbbbbbbbbbbb aaaaaaaa aaaaaaaa
360 370 380 390 400
d4iuja- ( 591 ) qIesmIeaessvkekdmTefFenspgveeg---------sigkvCrtlLA
d5d9aa3 ( 577 ) tVetiClresakaeenlvektlnekqmwigkkngeliaqpLreALrvqLV
aaaaaaaaaaaaa aaaaaaa
410 420 430 440 450
d4iuja- ( 643 ) kSVFnslYaspqLegFsaes-kLllivqald--------nlggly-aIe-
d5d9aa3 ( 627 ) qqFYfcIYndsqLegFcneqkkilmalegdkknkssfgfnpeglLekIee
aaaaaaa aaaaaaaaa aaaaaaa
460 470 480
d4iuja- ( 693 ) cLinDPWVLLnAswfnslth
d5d9aa3 ( 677 ) cLinNPMCLFmAqrlnelVieaSkrgakFfkt
aaaaaaaaaaa
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | 150 | 160 | 170 | 180 | 190 | 200 | 210 | 220 | 230 | 240 | 250 | 260 | 270 | 280 | 290 | 300 | 310 | 320 | 330 | 340 | 350 | 360 | 370 | 380 | 390 | 400 | 410 | 420 | 430 | 440 | 450 | 460 | 470 | 480 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | - | - | - | - | - | - | - | - | - | R | P | L | L | P | D | G | P | P | C | S | Q | R | S | K | F | L | L | M | D | A | L | K | L | S | I | E | D | P | S | H | G | E | G | I | P | L | Y | D | A | I | C | M | R | T | - | - | F | F | G | W | K | E | P | N | V | V | K | P | H | K | G | I | N | P | N | Y | L | L | S | W | K | Q | V | L | A | E | L | Q | D | I | E | N | I | P | R | T | K | N | M | K | K | T | S | Q | L | K | W | A | L | G | E | E | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | L | R | S | L | A | S | W | I | Q | N | E | F | N | K | A | C | E | L | T | D | S | - | S | W | I | E | - | - | - | - | - | L | D | E | I | G | E | D | A | A | P | - | - | - | - | - | I | H | I | A | S | M | R | R | N | Y | F | T | A | E | V | S | H | C | R | A | T | E | Y | I | M | K | G | V | Y | I | N | T | A | L | L | N | A | S | C | A | A | M | D | D | F | Q | L | I | P | M | I | S | K | C | R | T | E | - | - | - | - | - | - | - | - | - | G | R | R | K | T | N | L | Y | G | F | I | I | K | G | R | S | H | L | N | D | T | D | V | V | N | F | V | S | M | E | F | S | L | T | D | P | R | L | E | P | H | K | W | E | K | Y | C | V | L | E | V | G | D | M | L | L | R | S | A | I | G | H | V | S | R | P | M | F | L | Y | V | R | T | N | G | T | S | K | I | K | M | K | W | G | M | E | M | R | R | C | L | L | Q | S | L | Q | Q | I | E | S | M | I | E | A | E | S | S | V | K | E | K | D | M | T | E | F | F | E | N | S | P | G | V | E | E | G | - | - | - | - | - | - | - | - | - | S | I | G | K | V | C | R | T | L | L | A | K | S | V | F | N | S | L | Y | A | S | P | Q | L | E | G | F | S | A | E | S | - | K | L | L | L | I | V | Q | A | L | D | - | - | - | - | - | - | - | - | N | L | G | G | L | Y | - | A | I | E | - | C | L | I | N | D | P | W | V | L | L | N | A | S | W | F | N | S | L | T | H | - | - | - | - | - | - | - | - | - | - | - | - |
| I | K | H | Y | E | Q | N | K | F | R | Q | I | R | L | P | K | G | P | M | A | P | Y | T | H | K | F | L | M | E | E | A | W | M | F | T | K | I | S | D | P | E | R | S | R | A | G | E | I | L | I | D | F | F | K | K | G | N | L | S | A | I | R | P | K | D | K | P | L | Q | G | K | - | Y | P | I | H | Y | K | N | L | W | N | Q | I | K | A | A | I | A | D | - | - | - | - | - | - | R | T | M | V | I | N | E | N | D | H | S | E | F | L | G | G | R | A | S | K | K | I | P | E | I | S | L | T | Q | D | V | I | T | T | E | G | L | K | Q | S | E | N | K | L | P | E | P | R | S | F | P | R | W | F | N | A | E | W | M | W | A | I | K | D | S | D | L | T | G | W | V | P | M | A | E | Y | P | - | - | - | - | - | - | - | - | - | P | A | D | N | E | L | E | D | Y | A | E | H | L | N | K | T | M | E | G | V | L | Q | G | T | N | C | A | R | E | M | G | K | C | I | L | T | V | G | A | L | M | T | E | C | R | L | F | P | G | K | I | K | V | V | P | I | Y | A | R | S | K | E | R | K | S | M | Q | E | G | L | P | V | P | S | E | M | D | C | L | F | G | I | C | V | K | S | K | S | H | - | - | - | D | G | M | Y | T | I | I | T | F | E | F | S | I | R | E | P | - | - | N | L | E | K | H | Q | K | Y | T | V | F | E | A | G | H | T | T | V | R | M | - | - | K | K | R | E | V | P | L | Y | L | Y | C | R | T | T | A | L | S | K | I | K | N | D | W | L | S | K | A | R | R | C | F | I | T | T | M | D | T | V | E | T | I | C | L | R | E | S | A | K | A | E | E | N | L | V | E | K | T | L | N | E | K | Q | M | W | I | G | K | K | N | G | E | L | I | A | Q | P | L | R | E | A | L | R | V | Q | L | V | Q | Q | F | Y | F | C | I | Y | N | D | S | Q | L | E | G | F | C | N | E | Q | K | K | I | L | M | A | L | E | G | D | K | K | N | K | S | S | F | G | F | N | P | E | G | L | L | E | K | I | E | E | C | L | I | N | N | P | M | C | L | F | M | A | Q | R | L | N | E | L | V | I | E | A | S | K | R | G | A | K | F | F | K | T |
| Member | Chain | UniProtKB | GO term |
|---|