solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d3nwaa- ( 104 ) ikAentdanFyVÇppPtgatvvqfeqprrçptrpe---------------
d5mdmc- ( 1 ) ylsIAFPentkLdWKpVtkntry
d6tita- ( 1 ) kftivFPhnqkGnWknvpsnYhy
60 70 80 90 100
d3nwaa- ( 139 ) -----------gqnyteGIAVVFkeni---apykfkAtMyyKdVtvSVrf
d5mdmc- ( 24 ) ÇpmGgewflep--glqeesflssTPIgaTpsksdGflÇhAAkwVTtÇdfr
d6tita- ( 24 ) çPsSsDlnWhn--dligtalqvkmpksHkaiqAdGwMÇHASkWvttÇdfr
bbbbbbbbb bbbbbbbbbbbbb
110 120 130 140 150
d3nwaa- ( 176 ) grysqfmgifedrApVp---feEVidkInakGvÇrStAkyvrnnlettaf
d5mdmc- ( 72 ) wygpkyithsihnikPtrsdÇdtalasyksgtlvslgfPp-----esçgy
d6tita- ( 72 ) wygpkyithSirsftPsveqÇkeSieqtkqgtwlnpgfpp-----qsçgy
bbbb aaaaaa aa
160 170 180 190 200
d3nwaa- ( 224 ) hrddhetdmeLpanaatrtsrgWhTtdlkynPsrveafhryGtTVnÇiVe
d5mdmc- ( 117 ) ----------as----------------------------vtdseflVim
d6tita- ( 117 ) ----------at----------------------------vtdaeavivq
bbbbbbb
210 220 230 240 250
d3nwaa- ( 275 ) eVdArS-vypYd-eFvLatgdfVyMSPFyGyregshtehtsyaadrFkQv
d5mdmc- ( 129 ) ItphhVGvDDYrGhWVDp-----LFvggeçdq------------------
d6tita- ( 129 ) vtpHhvlvDEYtGewvDS-----qFingkçsn------------------
bbb bb
260 270 280 290 300
d3nwaa- ( 323 ) dgfyardlttkarataptTrNLLtTpkFTVAWdwvpkrpsVçTMtkwqeV
d5mdmc- ( 156 ) ------------------syÇdTihnssvWIPadqtkk--niçgqsFtpL
d6tita- ( 156 ) ------------------yiÇptvhnsttWHsdy-kvk--glçdsnlism
bb bbbb bbb
310 320 330 340 350
d3nwaa- ( 373 ) deMlRSeyg--------gsFrFsSdaisTTFtTnlt-EyplsrVdlGdçI
d5mdmc- ( 186 ) tVtVAYdktk--eIaagg-IvFkSk---YHshmeGarTçr---lsyçg--
d6tita- ( 185 ) ditFFSedgelsSlgkeg-tGFrsn---yFAYetGgkAçk---mqyçk--
bbbbbb bbbb bb
360 370 380 390 400
d3nwaa- ( 414 ) gkdArdAMdrIfarrYnathikVgqpQyYlAnggFLIAYQPLlsntlael
d5mdmc- ( 225 ) -------------------------rnGIkFpngEWVsLdv------ktr
d6tita- ( 226 ) -------------------------hwGvrlpsGvWFeMad------kd-
bbb bbb
410 420 430 440 450
d3nwaa- ( 464 ) yvehlrEqsrkppv---erIkttssiefArlqftynhiqrhvndmlgrva
d5mdmc- ( 244 ) IqekhLLpl-FkeÇpagtevr-S-Tlqsa-----qvlts----------e
d6tita- ( 244 ) ----lfaaArFpeÇpegssisAp-sqtsvd----vsliq----------d
460 470 480 490 500
d3nwaa- ( 526 ) iawçelqnheltlwnearkl---npnaiAsvtvg--rrVSArmlg----d
d5mdmc- ( 278 ) iQrildyslÇqnTWdkverkeplspldLSyLAskSPgkGLAYTvingtlS
d6tita- ( 275 ) verildyslÇqetWskiraglpisPvdlSylaPknPgtGPAFtiingtlk
aaaaaaaaaaaaaaaaaa aaaaa bbbbb
510 520 530 540 550
d3nwaa- ( 567 ) vMAvstÇvpVaadnViVqnsMrIs-----srpgaÇysrPlVsFrydqgpl
d5mdmc- ( 328 ) FAhtRYVrMwIdgpvlkepkGkrespsgi----ssdIWtqwfkygd----
d6tita- ( 325 ) YFEtRYiRvdiaapilsrMvGmi-sgttt----erelWddWapYed----
bbbbb bbb bb bbb bbb
560 570 580 590 600
d3nwaa- ( 613 ) veGQLgenneLrltrdaiepçtvgHrryFtFG--gGyVYFeeYayshQls
d5mdmc- ( 370 ) --meIGPNGLlkta------------ggykFpwhLigmelhelse
d6tita- ( 366 ) --veiGPnGvlrts------------sgyKFPlymig--------hg---
bbb 333 bb bbb
610 620 630 640 650
d3nwaa- ( 661 ) radittvstfidlnitmledhefvplevytrheikdsglldytevqrrnq
d5mdmc-
d6tita- ( 391 ) ------------mldsdlhlsskAqvFeHpHiqdAasqlpd---------
660 670
d3nwaa- ( 711 ) lhdlrfadidtvih
d5mdmc-
d6tita- ( 420 ) ---------deslffgdtglsk
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| Glycoprotein B | Glycoprotein B-like | Human herpesvirus 1 [TaxId: 10298] |  | view | |
| automated matches | automated matches | Chandipura virus [TaxId: 11272] | 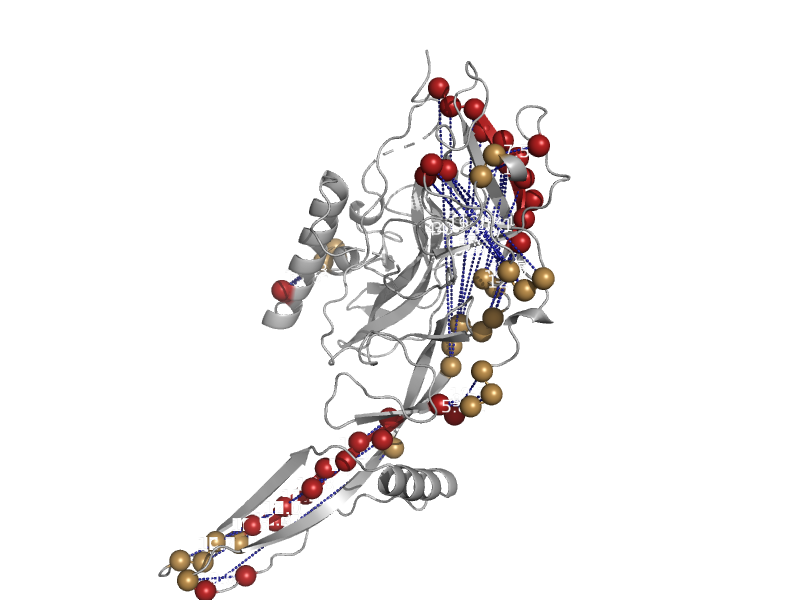 | view | |
| Spike glycoprotein | Spike glycoprotein-like | Recombinant vesicular stomatitis indiana virus rvsv-g/gfp [TaxId: 582817] | 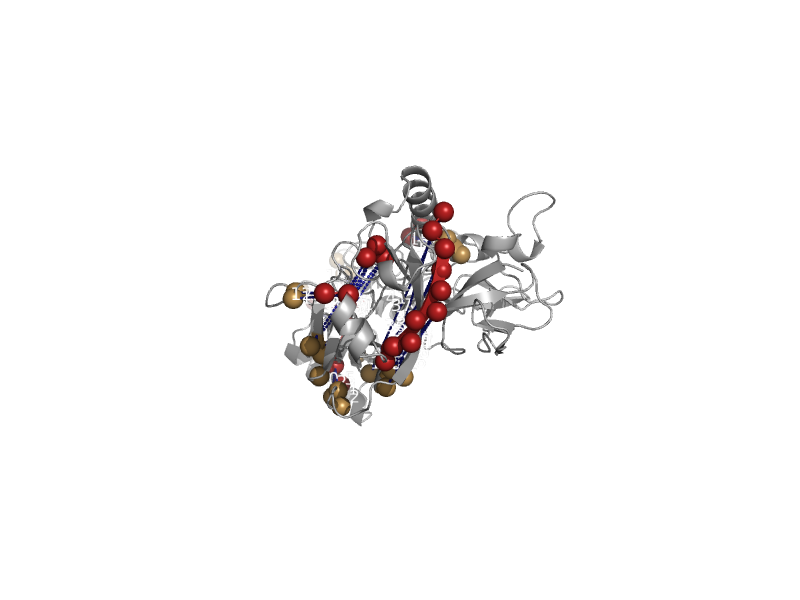 | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | 150 | 160 | 170 | 180 | 190 | 200 | 210 | 220 | 230 | 240 | 250 | 260 | 270 | 280 | 290 | 300 | 310 | 320 | 330 | 340 | 350 | 360 | 370 | 380 | 390 | 400 | 410 | 420 | 430 | 440 | 450 | 460 | 470 | 480 | 490 | 500 | 510 | 520 | 530 | 540 | 550 | 560 | 570 | 580 | 590 | 600 | 610 | 620 | 630 | 640 | 650 | 660 | 670 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I | K | A | E | N | T | D | A | N | F | Y | V | C | P | P | P | T | G | A | T | V | V | Q | F | E | Q | P | R | R | C | P | T | R | P | E | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | Q | N | Y | T | E | G | I | A | V | V | F | K | E | N | I | - | - | - | A | P | Y | K | F | K | A | T | M | Y | Y | K | D | V | T | V | S | V | R | F | G | R | Y | S | Q | F | M | G | I | F | E | D | R | A | P | V | P | - | - | - | F | E | E | V | I | D | K | I | N | A | K | G | V | C | R | S | T | A | K | Y | V | R | N | N | L | E | T | T | A | F | H | R | D | D | H | E | T | D | M | E | L | P | A | N | A | A | T | R | T | S | R | G | W | H | T | T | D | L | K | Y | N | P | S | R | V | E | A | F | H | R | Y | G | T | T | V | N | C | I | V | E | E | V | D | A | R | S | - | V | Y | P | Y | D | - | E | F | V | L | A | T | G | D | F | V | Y | M | S | P | F | Y | G | Y | R | E | G | S | H | T | E | H | T | S | Y | A | A | D | R | F | K | Q | V | D | G | F | Y | A | R | D | L | T | T | K | A | R | A | T | A | P | T | T | R | N | L | L | T | T | P | K | F | T | V | A | W | D | W | V | P | K | R | P | S | V | C | T | M | T | K | W | Q | E | V | D | E | M | L | R | S | E | Y | G | - | - | - | - | - | - | - | - | G | S | F | R | F | S | S | D | A | I | S | T | T | F | T | T | N | L | T | - | E | Y | P | L | S | R | V | D | L | G | D | C | I | G | K | D | A | R | D | A | M | D | R | I | F | A | R | R | Y | N | A | T | H | I | K | V | G | Q | P | Q | Y | Y | L | A | N | G | G | F | L | I | A | Y | Q | P | L | L | S | N | T | L | A | E | L | Y | V | E | H | L | R | E | Q | S | R | K | P | P | V | - | - | - | E | R | I | K | T | T | S | S | I | E | F | A | R | L | Q | F | T | Y | N | H | I | Q | R | H | V | N | D | M | L | G | R | V | A | I | A | W | C | E | L | Q | N | H | E | L | T | L | W | N | E | A | R | K | L | - | - | - | N | P | N | A | I | A | S | V | T | V | G | - | - | R | R | V | S | A | R | M | L | G | - | - | - | - | D | V | M | A | V | S | T | C | V | P | V | A | A | D | N | V | I | V | Q | N | S | M | R | I | S | - | - | - | - | - | S | R | P | G | A | C | Y | S | R | P | L | V | S | F | R | Y | D | Q | G | P | L | V | E | G | Q | L | G | E | N | N | E | L | R | L | T | R | D | A | I | E | P | C | T | V | G | H | R | R | Y | F | T | F | G | - | - | G | G | Y | V | Y | F | E | E | Y | A | Y | S | H | Q | L | S | R | A | D | I | T | T | V | S | T | F | I | D | L | N | I | T | M | L | E | D | H | E | F | V | P | L | E | V | Y | T | R | H | E | I | K | D | S | G | L | L | D | Y | T | E | V | Q | R | R | N | Q | L | H | D | L | R | F | A | D | I | D | T | V | I | H | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Y | L | S | I | A | F | P | E | N | T | K | L | D | W | K | P | V | T | K | N | T | R | Y | C | P | M | G | G | E | W | F | L | E | P | - | - | G | L | Q | E | E | S | F | L | S | S | T | P | I | G | A | T | P | S | K | S | D | G | F | L | C | H | A | A | K | W | V | T | T | C | D | F | R | W | Y | G | P | K | Y | I | T | H | S | I | H | N | I | K | P | T | R | S | D | C | D | T | A | L | A | S | Y | K | S | G | T | L | V | S | L | G | F | P | P | - | - | - | - | - | E | S | C | G | Y | - | - | - | - | - | - | - | - | - | - | A | S | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | V | T | D | S | E | F | L | V | I | M | I | T | P | H | H | V | G | V | D | D | Y | R | G | H | W | V | D | P | - | - | - | - | - | L | F | V | G | G | E | C | D | Q | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | S | Y | C | D | T | I | H | N | S | S | V | W | I | P | A | D | Q | T | K | K | - | - | N | I | C | G | Q | S | F | T | P | L | T | V | T | V | A | Y | D | K | T | K | - | - | E | I | A | A | G | G | - | I | V | F | K | S | K | - | - | - | Y | H | S | H | M | E | G | A | R | T | C | R | - | - | - | L | S | Y | C | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | R | N | G | I | K | F | P | N | G | E | W | V | S | L | D | V | - | - | - | - | - | - | K | T | R | I | Q | E | K | H | L | L | P | L | - | F | K | E | C | P | A | G | T | E | V | R | - | S | - | T | L | Q | S | A | - | - | - | - | - | Q | V | L | T | S | - | - | - | - | - | - | - | - | - | - | E | I | Q | R | I | L | D | Y | S | L | C | Q | N | T | W | D | K | V | E | R | K | E | P | L | S | P | L | D | L | S | Y | L | A | S | K | S | P | G | K | G | L | A | Y | T | V | I | N | G | T | L | S | F | A | H | T | R | Y | V | R | M | W | I | D | G | P | V | L | K | E | P | K | G | K | R | E | S | P | S | G | I | - | - | - | - | S | S | D | I | W | T | Q | W | F | K | Y | G | D | - | - | - | - | - | - | M | E | I | G | P | N | G | L | L | K | T | A | - | - | - | - | - | - | - | - | - | - | - | - | G | G | Y | K | F | P | W | H | L | I | G | M | E | L | H | E | L | S | E | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | K | F | T | I | V | F | P | H | N | Q | K | G | N | W | K | N | V | P | S | N | Y | H | Y | C | P | S | S | S | D | L | N | W | H | N | - | - | D | L | I | G | T | A | L | Q | V | K | M | P | K | S | H | K | A | I | Q | A | D | G | W | M | C | H | A | S | K | W | V | T | T | C | D | F | R | W | Y | G | P | K | Y | I | T | H | S | I | R | S | F | T | P | S | V | E | Q | C | K | E | S | I | E | Q | T | K | Q | G | T | W | L | N | P | G | F | P | P | - | - | - | - | - | Q | S | C | G | Y | - | - | - | - | - | - | - | - | - | - | A | T | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | V | T | D | A | E | A | V | I | V | Q | V | T | P | H | H | V | L | V | D | E | Y | T | G | E | W | V | D | S | - | - | - | - | - | Q | F | I | N | G | K | C | S | N | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Y | I | C | P | T | V | H | N | S | T | T | W | H | S | D | Y | - | K | V | K | - | - | G | L | C | D | S | N | L | I | S | M | D | I | T | F | F | S | E | D | G | E | L | S | S | L | G | K | E | G | - | T | G | F | R | S | N | - | - | - | Y | F | A | Y | E | T | G | G | K | A | C | K | - | - | - | M | Q | Y | C | K | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | H | W | G | V | R | L | P | S | G | V | W | F | E | M | A | D | - | - | - | - | - | - | K | D | - | - | - | - | - | L | F | A | A | A | R | F | P | E | C | P | E | G | S | S | I | S | A | P | - | S | Q | T | S | V | D | - | - | - | - | V | S | L | I | Q | - | - | - | - | - | - | - | - | - | - | D | V | E | R | I | L | D | Y | S | L | C | Q | E | T | W | S | K | I | R | A | G | L | P | I | S | P | V | D | L | S | Y | L | A | P | K | N | P | G | T | G | P | A | F | T | I | I | N | G | T | L | K | Y | F | E | T | R | Y | I | R | V | D | I | A | A | P | I | L | S | R | M | V | G | M | I | - | S | G | T | T | T | - | - | - | - | E | R | E | L | W | D | D | W | A | P | Y | E | D | - | - | - | - | - | - | V | E | I | G | P | N | G | V | L | R | T | S | - | - | - | - | - | - | - | - | - | - | - | - | S | G | Y | K | F | P | L | Y | M | I | G | - | - | - | - | - | - | - | - | H | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | M | L | D | S | D | L | H | L | S | S | K | A | Q | V | F | E | H | P | H | I | Q | D | A | A | S | Q | L | P | D | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | D | E | S | L | F | F | G | D | T | G | L | S | K |
| Member | Chain | UniProtKB | GO term |
|---|
