solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d2dm9a1 ( 81 ) eiissvleevkrrletmsedeyFesVKalLkeAIkeLnekkVrVMSnekT
d3k5ba1 ( 78 ) evleevrrrvreaealpq-pewpvraaap------------gaavan--p
d4efae2 ( 98 ) gifeetkeklsgiAnn--rdeYkpiLqslIVeALlkLlepkAiVkALerD
aaaaaaaaaaa
60 70 80 90 100
d2dm9a1 ( 131 ) LglIasrieeIkselg------dV-sIelg-etvd---TmGGViVeTedg
d3k5ba1 ( 128 ) lphaag-----------------------------vlaeplgvavg-aeg
d4efae2 ( 146 ) vdlIesMkddImreYgekaqrapLeeIvIsndylnkdlvsGGVVVsNasd
110 120 130
d2dm9a1 ( 170 ) riridNtfearmerfegeirstiakvlfg
d3k5ba1 ( 161 ) tqvnslardrawdas--svaqalwg
d4efae2 ( 196 ) kieinNTleerlkllseealpairlelygps
aaaa
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| V-type ATPase subunit E C-terminal domain | V-type ATPase subunit E C-terminal domain | Pyrococcus horikoshii [TaxId: 53953] | 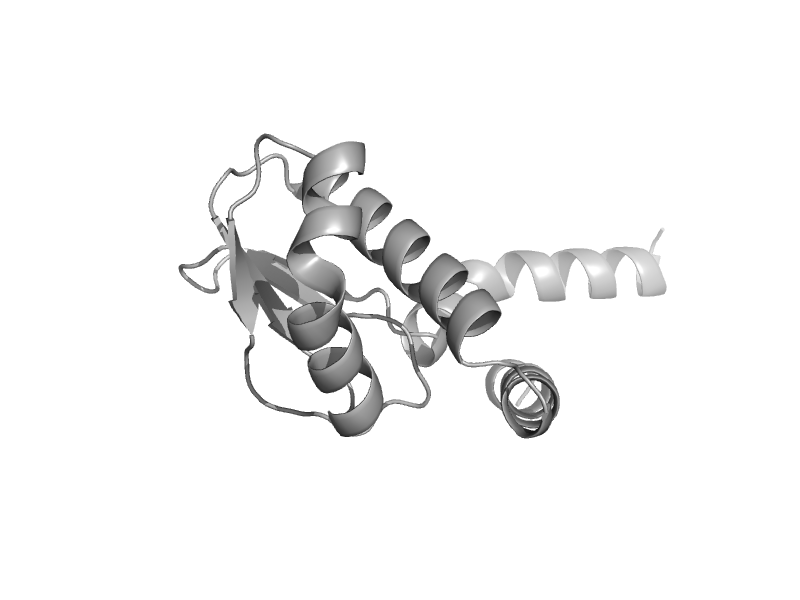 | view | |
| V-type ATPase subunit E C-terminal domain | V-type ATPase subunit E C-terminal domain | Thermus thermophilus HB8 [TaxId: 300852] | 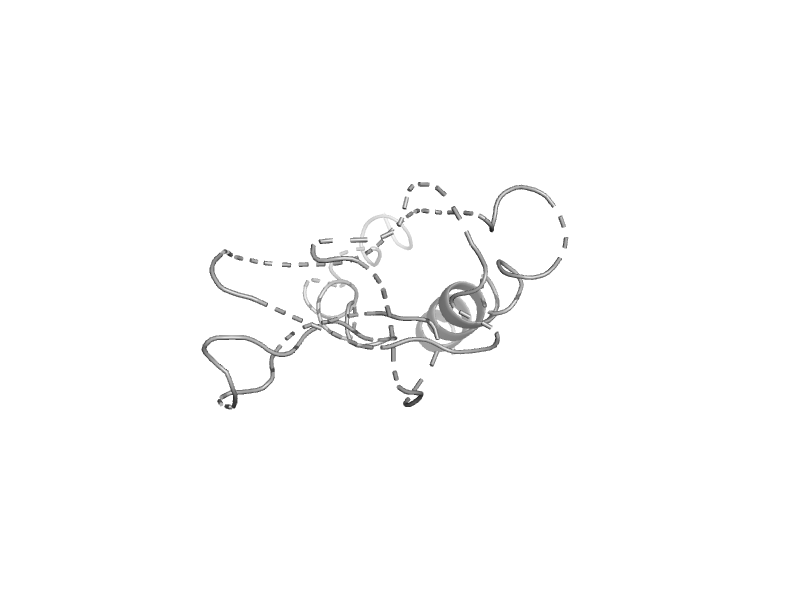 | view | |
| V-type ATPase subunit E C-terminal domain | V-type ATPase subunit E C-terminal domain | Baker's yeast (Saccharomyces cerevisiae) [TaxId: 4932] | 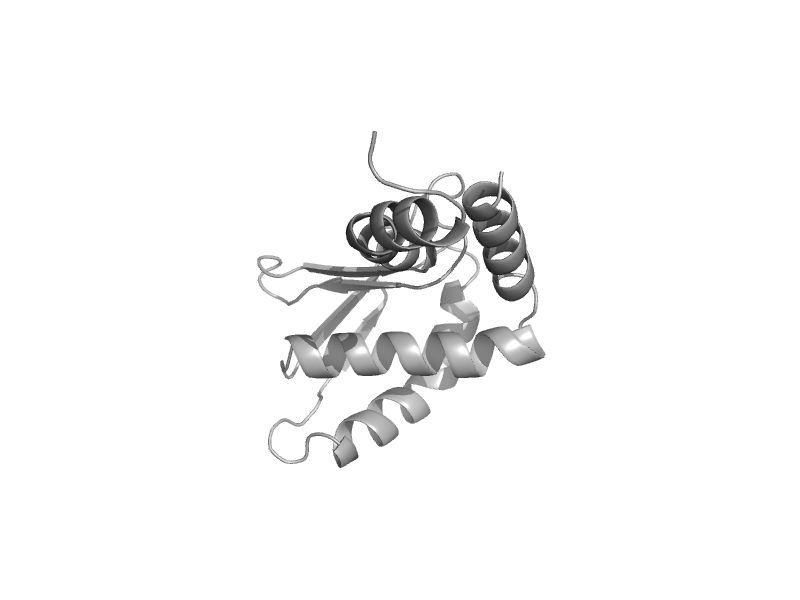 | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E | I | I | S | S | V | L | E | E | V | K | R | R | L | E | T | M | S | E | D | E | Y | F | E | S | V | K | A | L | L | K | E | A | I | K | E | L | N | E | K | K | V | R | V | M | S | N | E | K | T | L | G | L | I | A | S | R | I | E | E | I | K | S | E | L | G | - | - | - | - | - | - | D | V | - | S | I | E | L | G | - | E | T | V | D | - | - | - | T | M | G | G | V | I | V | E | T | E | D | G | R | I | R | I | D | N | T | F | E | A | R | M | E | R | F | E | G | E | I | R | S | T | I | A | K | V | L | F | G | - | - |
| E | V | L | E | E | V | R | R | R | V | R | E | A | E | A | L | P | Q | - | P | E | W | P | V | R | A | A | A | P | - | - | - | - | - | - | - | - | - | - | - | - | G | A | A | V | A | N | - | - | P | L | P | H | A | A | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | V | L | A | E | P | L | G | V | A | V | G | - | A | E | G | T | Q | V | N | S | L | A | R | D | R | A | W | D | A | S | - | - | S | V | A | Q | A | L | W | G | - | - | - | - | - | - |
| G | I | F | E | E | T | K | E | K | L | S | G | I | A | N | N | - | - | R | D | E | Y | K | P | I | L | Q | S | L | I | V | E | A | L | L | K | L | L | E | P | K | A | I | V | K | A | L | E | R | D | V | D | L | I | E | S | M | K | D | D | I | M | R | E | Y | G | E | K | A | Q | R | A | P | L | E | E | I | V | I | S | N | D | Y | L | N | K | D | L | V | S | G | G | V | V | V | S | N | A | S | D | K | I | E | I | N | N | T | L | E | E | R | L | K | L | L | S | E | E | A | L | P | A | I | R | L | E | L | Y | G | P | S |
| Member | Chain | UniProtKB | GO term |
|---|---|---|---|
| d2dm9a1 | A | O57724 | GO:0005524 |
| d2dm9a1 | A | O57724 | GO:0006754 |
| d2dm9a1 | A | O57724 | GO:0033178 |
| d2dm9a1 | A | O57724 | GO:0042777 |
| d2dm9a1 | A | O57724 | GO:0046933 |
| d2dm9a1 | A | O57724 | GO:0046961 |
| d2dm9a1 | A | O57724 | GO:1902600 |
