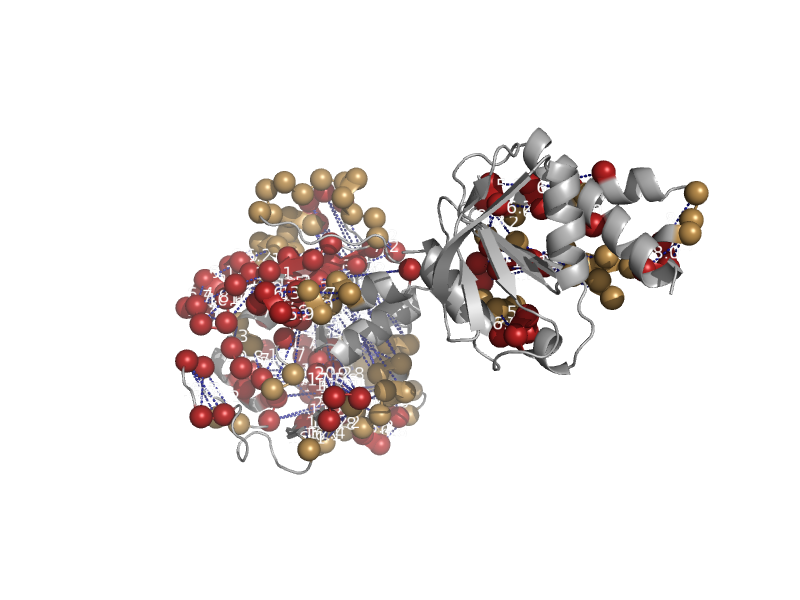
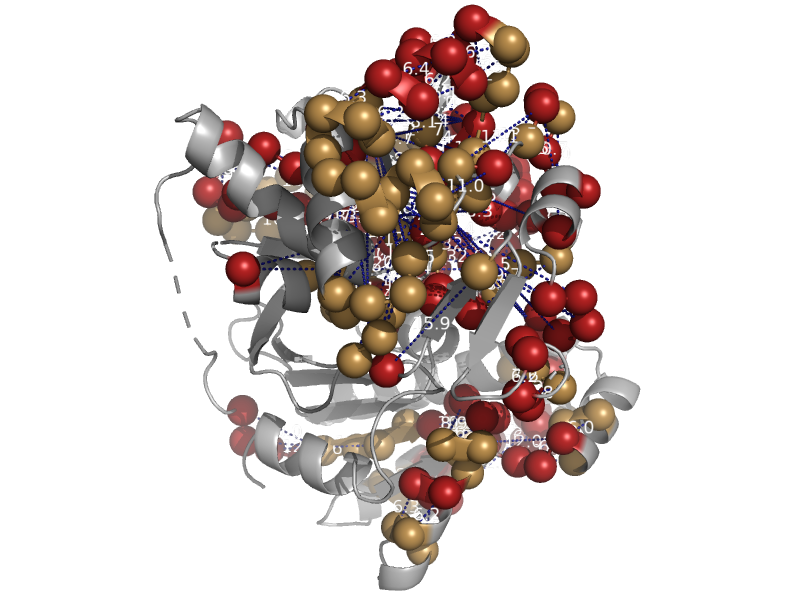
solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d4xfma- ( 9 ) gqvlvvADdftGAndAgvglaqhgArvsvvfdv--ntlhadll-gdAvv
d5dmha- ( 9 ) rPllGCiADDFtgAtDlAntlvrng-rtvqtiglpdvgavqdigeAdAlv
bbbbb aaaaaaaaaaaaa bbb bbb
60 70 80 90 100
d4xfma- ( 55 ) intdSraarddvAsqrtaaAvaaWqavgGkGWiiKkidstlr----GnlG
d5dmha- ( 59 ) vAlkSrtipaveAvaqSlaAlqwlraqgCr-qFvFKYcStFdStdaGniG
bb aaaaaaaaaaaaaaaaa bbbbb aa
110 120 130 140 150
d4xfma- ( 101 ) aEvaaAlsaAdvpvAliAAASPtlgrvtrqGevwvngrrltdtefAsdpk
d5dmha- ( 108 ) pvaeallaaldsdFtiACPAFpengRtifrGhlfvgdallnesg-ehhpl
aaaaaaaaaa bb 333 bbbb bbbb bb333
160 170 180 190 200
d4xfma- ( 151 ) tpvtsAsiaaRlaeqtalpvaeihldevrq--anlahrlqqlAdegtrli
d5dmha- ( 158 ) tp-tdAslvrvlqrqSknkvgllrydAvargahataeriaalrsdgvr-A
aaaaaa bb aaaa aaaaaaaaaa
210 220 230 240 250
d4xfma- ( 199 ) ilDtdvqdDlthivnaAralpfrPllvGSaglSdAlAtaq-----dftrk
d5dmha- ( 208 ) iADAvsdadlftlGeACan---lplitGGSGiAlGlPenfrragllpqrs
bb aaaaaaaaaa bbbb aaaaa
260 270 280 290 300
d4xfma- ( 244 ) te---kPllAvvGsmsdiAqkqiaAArlrsDvtlveidinalfspdsstv
d5dmha- ( 259 ) vpaidgpGvvlAGsasratngqvarWle-qgrpAlridplalAr------
bbbbb aaaaaaaaaaa bbb
310 320 330 340 350
d4xfma- ( 291 ) masqcedAlkaltn--ghHCiirtcq--------------qlgetishyl
d5dmha- ( 302 ) geavadaAlafAaghgepvliyAtsspdevkavqaelgverAghlveqCl
aaaaaaaaaa bbb aaaaaaaaaa
360 370 380 390 400
d4xfma- ( 344 ) GeltrsivqallPgGlYlSggdiaiavatalgAtgfqikgqiascvpwGy
d5dmha- ( 352 ) atvAagllar-gtrRFvvAgGetSGavvqalgvrAlriGaqiapGvPAtv
aaaaaaaaa bbbbb aaaaaaaaaa bbbbbbbb bbbbb
410 420 430
d4xfma- ( 406 ) llnSivgmtpvMtKaggfgnettlldvlrfiee
d5dmha- ( 401 ) tl-da-kplAlAlKsgnFGgpdFFdeAlrqlGgh
b bbbbb aaaaaaaaa
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | 150 | 160 | 170 | 180 | 190 | 200 | 210 | 220 | 230 | 240 | 250 | 260 | 270 | 280 | 290 | 300 | 310 | 320 | 330 | 340 | 350 | 360 | 370 | 380 | 390 | 400 | 410 | 420 | 430 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | G | Q | V | L | V | V | A | D | D | F | T | G | A | N | D | A | G | V | G | L | A | Q | H | G | A | R | V | S | V | V | F | D | V | - | - | N | T | L | H | A | D | L | L | - | G | D | A | V | V | I | N | T | D | S | R | A | A | R | D | D | V | A | S | Q | R | T | A | A | A | V | A | A | W | Q | A | V | G | G | K | G | W | I | I | K | K | I | D | S | T | L | R | - | - | - | - | G | N | L | G | A | E | V | A | A | A | L | S | A | A | D | V | P | V | A | L | I | A | A | A | S | P | T | L | G | R | V | T | R | Q | G | E | V | W | V | N | G | R | R | L | T | D | T | E | F | A | S | D | P | K | T | P | V | T | S | A | S | I | A | A | R | L | A | E | Q | T | A | L | P | V | A | E | I | H | L | D | E | V | R | Q | - | - | A | N | L | A | H | R | L | Q | Q | L | A | D | E | G | T | R | L | I | I | L | D | T | D | V | Q | D | D | L | T | H | I | V | N | A | A | R | A | L | P | F | R | P | L | L | V | G | S | A | G | L | S | D | A | L | A | T | A | Q | - | - | - | - | - | D | F | T | R | K | T | E | - | - | - | K | P | L | L | A | V | V | G | S | M | S | D | I | A | Q | K | Q | I | A | A | A | R | L | R | S | D | V | T | L | V | E | I | D | I | N | A | L | F | S | P | D | S | S | T | V | M | A | S | Q | C | E | D | A | L | K | A | L | T | N | - | - | G | H | H | C | I | I | R | T | C | Q | - | - | - | - | - | - | - | - | - | - | - | - | - | - | Q | L | G | E | T | I | S | H | Y | L | G | E | L | T | R | S | I | V | Q | A | L | L | P | G | G | L | Y | L | S | G | G | D | I | A | I | A | V | A | T | A | L | G | A | T | G | F | Q | I | K | G | Q | I | A | S | C | V | P | W | G | Y | L | L | N | S | I | V | G | M | T | P | V | M | T | K | A | G | G | F | G | N | E | T | T | L | L | D | V | L | R | F | I | E | E | - |
| R | P | L | L | G | C | I | A | D | D | F | T | G | A | T | D | L | A | N | T | L | V | R | N | G | - | R | T | V | Q | T | I | G | L | P | D | V | G | A | V | Q | D | I | G | E | A | D | A | L | V | V | A | L | K | S | R | T | I | P | A | V | E | A | V | A | Q | S | L | A | A | L | Q | W | L | R | A | Q | G | C | R | - | Q | F | V | F | K | Y | C | S | T | F | D | S | T | D | A | G | N | I | G | P | V | A | E | A | L | L | A | A | L | D | S | D | F | T | I | A | C | P | A | F | P | E | N | G | R | T | I | F | R | G | H | L | F | V | G | D | A | L | L | N | E | S | G | - | E | H | H | P | L | T | P | - | T | D | A | S | L | V | R | V | L | Q | R | Q | S | K | N | K | V | G | L | L | R | Y | D | A | V | A | R | G | A | H | A | T | A | E | R | I | A | A | L | R | S | D | G | V | R | - | A | I | A | D | A | V | S | D | A | D | L | F | T | L | G | E | A | C | A | N | - | - | - | L | P | L | I | T | G | G | S | G | I | A | L | G | L | P | E | N | F | R | R | A | G | L | L | P | Q | R | S | V | P | A | I | D | G | P | G | V | V | L | A | G | S | A | S | R | A | T | N | G | Q | V | A | R | W | L | E | - | Q | G | R | P | A | L | R | I | D | P | L | A | L | A | R | - | - | - | - | - | - | G | E | A | V | A | D | A | A | L | A | F | A | A | G | H | G | E | P | V | L | I | Y | A | T | S | S | P | D | E | V | K | A | V | Q | A | E | L | G | V | E | R | A | G | H | L | V | E | Q | C | L | A | T | V | A | A | G | L | L | A | R | - | G | T | R | R | F | V | V | A | G | G | E | T | S | G | A | V | V | Q | A | L | G | V | R | A | L | R | I | G | A | Q | I | A | P | G | V | P | A | T | V | T | L | - | D | A | - | K | P | L | A | L | A | L | K | S | G | N | F | G | G | P | D | F | F | D | E | A | L | R | Q | L | G | G | H |
| Member | Chain | UniProtKB | GO term |
|---|