solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1yu0a1 ( 5 ) vqfrggttaqhatftGaareiTvDtdkntvVvHdgatagGfpla------
d1zrua3 ( 2 ) tiknftfgsnndGk
60 70 80 90 100
d1yu0a1 ( 49 ) -rhdlvk-----tafikadksavAFtrtgnatAsIkagTiVeVngklvqF
d1zrua3 ( 26 ) lymmltgMdyrtIrrkdws----splnta-lnVqYt-nTsIiAGGRyFeL
bb bb bbbb bbbbb bbbbb
110 120 130 140 150
d1yu0a1 ( 93 ) tadtaItmpaLtagtdYAIYVCdd-------gtVrAdsnfs----aPtgy
d1zrua3 ( 70 ) l-netvaLk---gdsvNyIHAnIdltqtanPVsLsAetannsnGvdINng
bb bbbbbbb bbbb
160 170 180 190
d1yu0a1 ( 132 ) tsttArkVGGFhyapgsnaaaqaggnttaqIneysLwdi
d1zrua3 ( 116 ) sgvLkVCFDiVtTsg-------------tgVt---stkpiv
bbbbbbbbb b bb
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| Major tropism determinant (Mtd), N-terminal domain | Mtd domain-like | Bordetella phage bpp-1 [TaxId: 194699] | 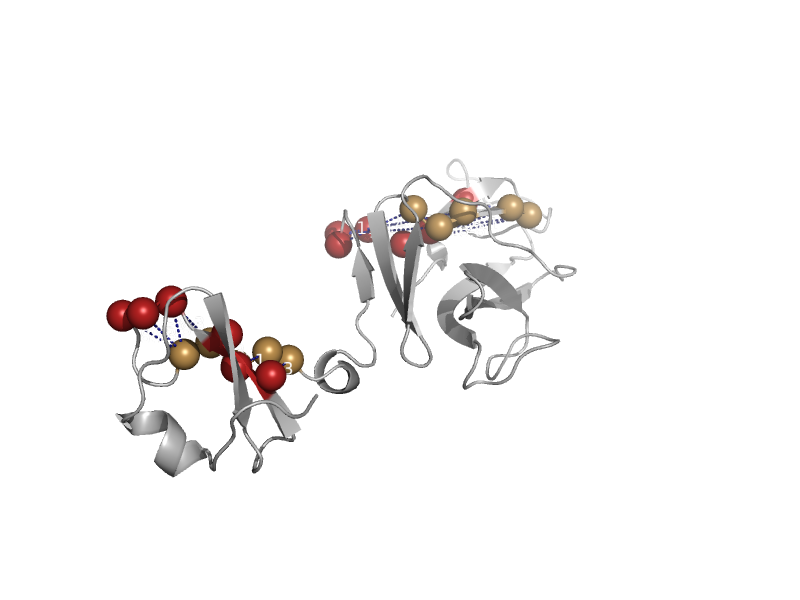 | view | |
| Receptor binding protein, rbp, N-terminal domain | Lactophage receptor-binding protein N-terminal domain | Lactococcus lactis phage p2 [TaxId: 100641] | 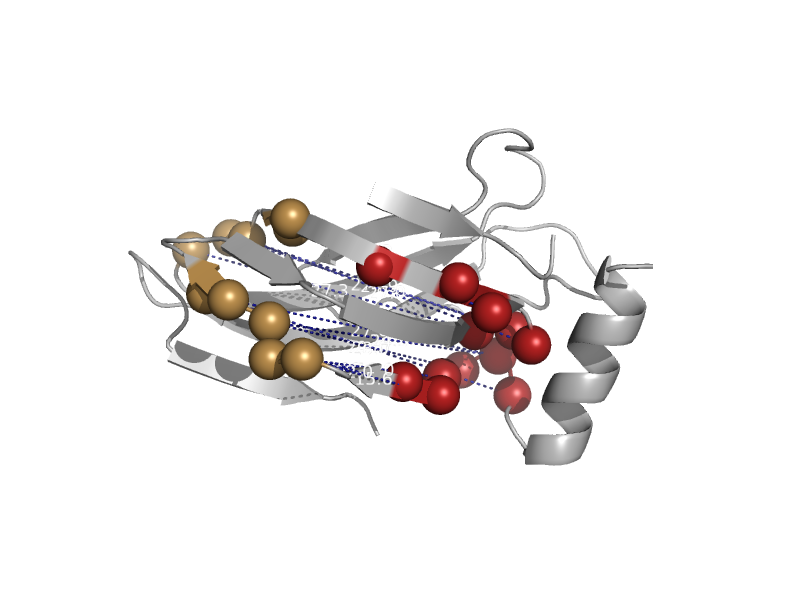 | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | 150 | 160 | 170 | 180 | 190 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| V | Q | F | R | G | G | T | T | A | Q | H | A | T | F | T | G | A | A | R | E | I | T | V | D | T | D | K | N | T | V | V | V | H | D | G | A | T | A | G | G | F | P | L | A | - | - | - | - | - | - | - | R | H | D | L | V | K | - | - | - | - | - | T | A | F | I | K | A | D | K | S | A | V | A | F | T | R | T | G | N | A | T | A | S | I | K | A | G | T | I | V | E | V | N | G | K | L | V | Q | F | T | A | D | T | A | I | T | M | P | A | L | T | A | G | T | D | Y | A | I | Y | V | C | D | D | - | - | - | - | - | - | - | G | T | V | R | A | D | S | N | F | S | - | - | - | - | A | P | T | G | Y | T | S | T | T | A | R | K | V | G | G | F | H | Y | A | P | G | S | N | A | A | A | Q | A | G | G | N | T | T | A | Q | I | N | E | Y | S | L | W | D | I | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | T | I | K | N | F | T | F | G | S | N | N | D | G | K | L | Y | M | M | L | T | G | M | D | Y | R | T | I | R | R | K | D | W | S | - | - | - | - | S | P | L | N | T | A | - | L | N | V | Q | Y | T | - | N | T | S | I | I | A | G | G | R | Y | F | E | L | L | - | N | E | T | V | A | L | K | - | - | - | G | D | S | V | N | Y | I | H | A | N | I | D | L | T | Q | T | A | N | P | V | S | L | S | A | E | T | A | N | N | S | N | G | V | D | I | N | N | G | S | G | V | L | K | V | C | F | D | I | V | T | T | S | G | - | - | - | - | - | - | - | - | - | - | - | - | - | T | G | V | T | - | - | - | S | T | K | P | I | V |
| Member | Chain | UniProtKB | GO term |
|---|---|---|---|
| d1yu0a1 | A | Q775D6 | GO:0005515 |
| d1yu0a1 | A | Q775D6 | GO:0046718 |
| d1yu0a1 | A | Q775D6 | GO:0098015 |
| d1yu0a1 | A | Q775D6 | GO:0098024 |
| d1yu0a1 | A | Q775D6 | GO:0098671 |
| d1yu0a1 | A | Q775D6 | GO:0098678 |
| d1zrua3 | A | Q71AW2 | GO:0007155 |
| d1zrua3 | A | Q71AW2 | GO:0019058 |
| d1zrua3 | A | Q71AW2 | GO:0019062 |
| d1zrua3 | A | Q71AW2 | GO:0046718 |
| d1zrua3 | A | Q71AW2 | GO:0098015 |
| d1zrua3 | A | Q71AW2 | GO:0098025 |
| d1zrua3 | A | Q71AW2 | GO:0098670 |
| d1zrua3 | A | Q71AW2 | GO:0007155 |
| d1zrua3 | A | Q71AW2 | GO:0019058 |
| d1zrua3 | A | Q71AW2 | GO:0019062 |
| d1zrua3 | A | Q71AW2 | GO:0046718 |
| d1zrua3 | A | Q71AW2 | GO:0098015 |
| d1zrua3 | A | Q71AW2 | GO:0098025 |
| d1zrua3 | A | Q71AW2 | GO:0098670 |
