solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d2f2ha1 ( 666 ) ntLlalgnndqrpdyvwhegTaFhLfnLqdg--heAvceVpaa-------
d2g3ma4 ( 605 ) gsIipl-----------egdeLiVyg--------etsfkr----------
d2xvla5 ( 871 ) gaIvptgpliqhvd-eglnsplLITVyTg--angsfdiyeddgrsl-kyq
d3l4ya4 ( 732 ) gyIfptqqpntttl-asrknpLgLiIaLdenkeAkgelfwddgetkdtva
d3weoa4 ( 757 ) gnivAmqgeamttq-aarstpFhllvvMsdhvaStgelfldngiemdigg
bbbb bbbbb bbbbbb
60 70 80 90 100
d2f2ha1 ( 707 ) dgsviFtLkAart----gntItVtgag---------eAknwtLCLrnvv-
d2g3ma4 ( 626 ) ydn--AeItSs------sneIkFs-----------reiyVskLtIts-ek
d2xvla5 ( 917 ) -qgewSrIpLsYddv--tgtLiIgdrvgsFtgmad--eRnIrVrFiag--
d3l4ya4 ( 781 ) -nkvyLlCeFsVtq----nrLeVn-isqs-tykDpnnLaFneIkIlgTee
d3weoa4 ( 806 ) pggkwtlvrFfAesginnltisSe-vv-nrgyAmsqrwvMdkitilglkr
bbbbbbb bbbb b bbbbbbbb
110 120 130 140
d2f2ha1 ( 743 ) kVngLq-----dGsqaese------qGLvVkpq---gnaLtItl
d2g3ma4 ( 656 ) pVskIiVddskeiqVektm-----qntYvAkInqkIrgkInle
d2xvla5 ( 960 ) ----------------ptadatnfdkAaAeavtYt-gksvsIkrpr
d3l4ya4 ( 824 ) PsnVtVkhngvpstsPtvtydsnlkvAiItdIdLlLgeaytVew
d3weoa4 ( 854 ) rvrik-----------------gfvvsvisdlrqlvGqafkleleFe
bb b bbb
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| Putative glucosidase YicI, C-terminal domain | Glycolsyl hydrolase family 31 C-terminal domain | Escherichia coli [TaxId: 562] | 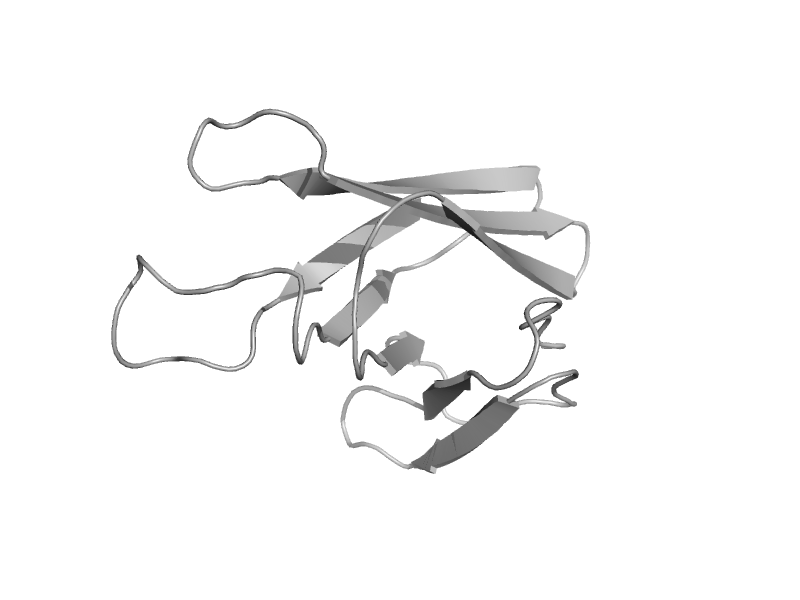 | view | |
| Maltase, C-terminal domain | Glycolsyl hydrolase family 31 C-terminal domain | Sulfolobus solfataricus [TaxId: 2287] | 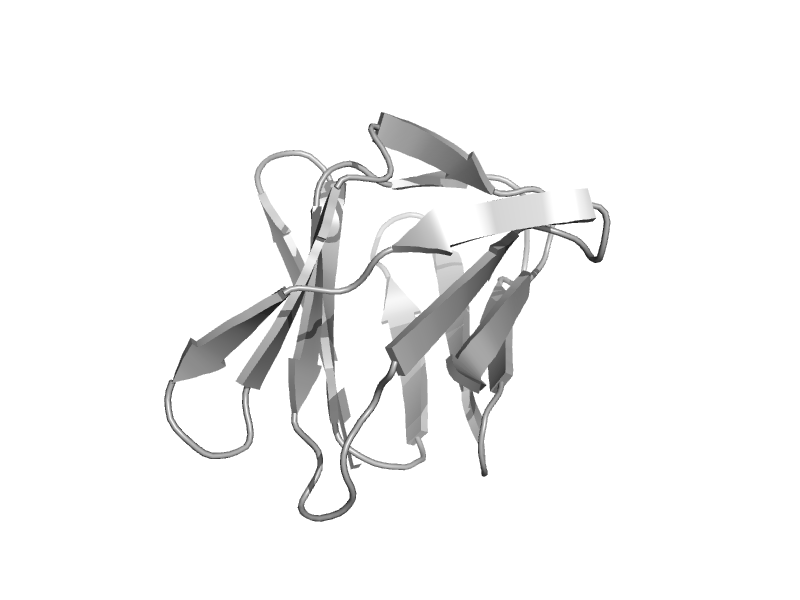 | view | |
| Alpha-xylosidase, C-terminal domain | Glycolsyl hydrolase family 31 C-terminal domain | Pseudomonas cellulosa (Cellvibrio japonicus) [TaxId: 155077] | 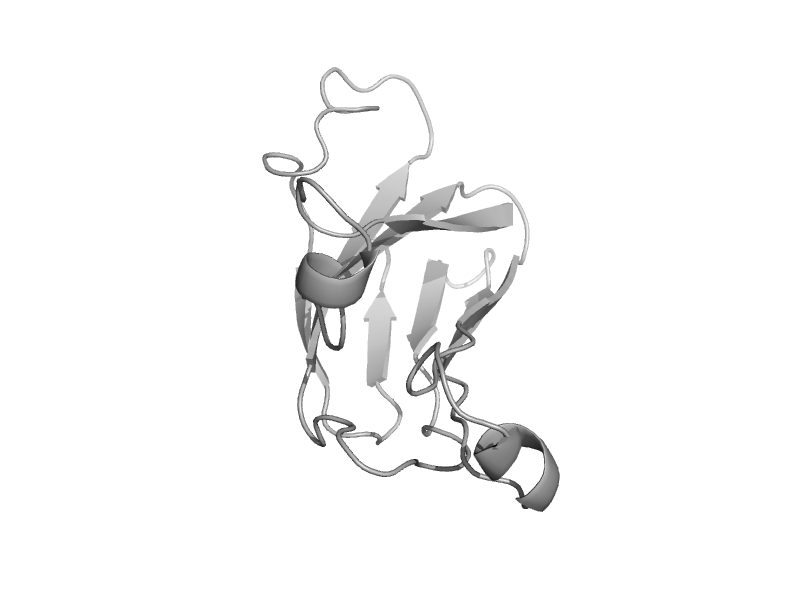 | view | |
| automated matches | automated matches | Human (Homo sapiens) [TaxId: 9606] | 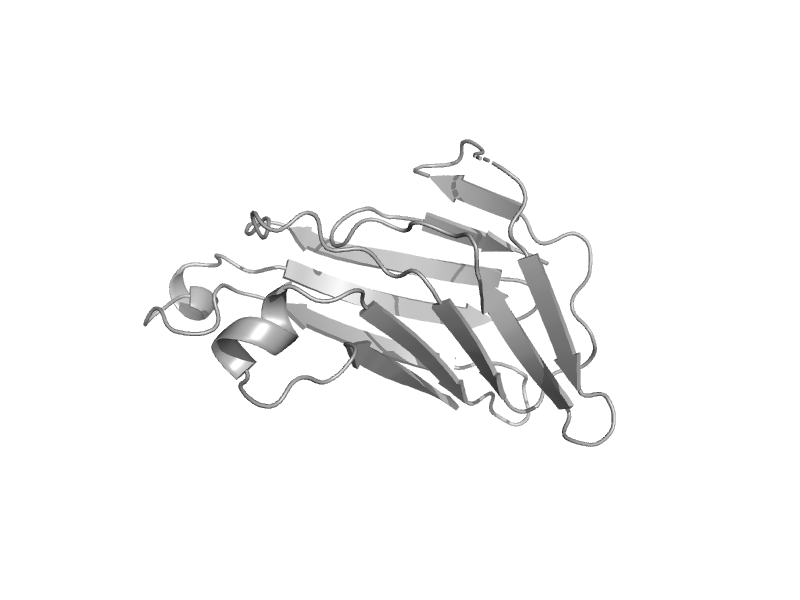 | view | |
| Maltase, C-terminal domain | Glycolsyl hydrolase family 31 C-terminal domain | Beta vulgaris [TaxId: 161934] |  | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | T | L | L | A | L | G | N | N | D | Q | R | P | D | Y | V | W | H | E | G | T | A | F | H | L | F | N | L | Q | D | G | - | - | H | E | A | V | C | E | V | P | A | A | - | - | - | - | - | - | - | D | G | S | V | I | F | T | L | K | A | A | R | T | - | - | - | - | G | N | T | I | T | V | T | G | A | G | - | - | - | - | - | - | - | - | - | E | A | K | N | W | T | L | C | L | R | N | V | V | - | K | V | N | G | L | Q | - | - | - | - | - | D | G | S | Q | A | E | S | E | - | - | - | - | - | - | Q | G | L | V | V | K | P | Q | - | - | - | G | N | A | L | T | I | T | L | - | - | - |
| G | S | I | I | P | L | - | - | - | - | - | - | - | - | - | - | - | E | G | D | E | L | I | V | Y | G | - | - | - | - | - | - | - | - | E | T | S | F | K | R | - | - | - | - | - | - | - | - | - | - | Y | D | N | - | - | A | E | I | T | S | S | - | - | - | - | - | - | S | N | E | I | K | F | S | - | - | - | - | - | - | - | - | - | - | - | R | E | I | Y | V | S | K | L | T | I | T | S | - | E | K | P | V | S | K | I | I | V | D | D | S | K | E | I | Q | V | E | K | T | M | - | - | - | - | - | Q | N | T | Y | V | A | K | I | N | Q | K | I | R | G | K | I | N | L | E | - | - | - | - |
| G | A | I | V | P | T | G | P | L | I | Q | H | V | D | - | E | G | L | N | S | P | L | L | I | T | V | Y | T | G | - | - | A | N | G | S | F | D | I | Y | E | D | D | G | R | S | L | - | K | Y | Q | - | Q | G | E | W | S | R | I | P | L | S | Y | D | D | V | - | - | T | G | T | L | I | I | G | D | R | V | G | S | F | T | G | M | A | D | - | - | E | R | N | I | R | V | R | F | I | A | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | P | T | A | D | A | T | N | F | D | K | A | A | A | E | A | V | T | Y | T | - | G | K | S | V | S | I | K | R | P | R | - |
| G | Y | I | F | P | T | Q | Q | P | N | T | T | T | L | - | A | S | R | K | N | P | L | G | L | I | I | A | L | D | E | N | K | E | A | K | G | E | L | F | W | D | D | G | E | T | K | D | T | V | A | - | N | K | V | Y | L | L | C | E | F | S | V | T | Q | - | - | - | - | N | R | L | E | V | N | - | I | S | Q | S | - | T | Y | K | D | P | N | N | L | A | F | N | E | I | K | I | L | G | T | E | E | P | S | N | V | T | V | K | H | N | G | V | P | S | T | S | P | T | V | T | Y | D | S | N | L | K | V | A | I | I | T | D | I | D | L | L | L | G | E | A | Y | T | V | E | W | - | - | - |
| G | N | I | V | A | M | Q | G | E | A | M | T | T | Q | - | A | A | R | S | T | P | F | H | L | L | V | V | M | S | D | H | V | A | S | T | G | E | L | F | L | D | N | G | I | E | M | D | I | G | G | P | G | G | K | W | T | L | V | R | F | F | A | E | S | G | I | N | N | L | T | I | S | S | E | - | V | V | - | N | R | G | Y | A | M | S | Q | R | W | V | M | D | K | I | T | I | L | G | L | K | R | R | V | R | I | K | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | F | V | V | S | V | I | S | D | L | R | Q | L | V | G | Q | A | F | K | L | E | L | E | F | E |
| Member | Chain | UniProtKB | GO term |
|---|---|---|---|
| d2f2ha1 | A | P31434 | GO:0003824 |
| d2f2ha1 | A | P31434 | GO:0004553 |
| d2f2ha1 | A | P31434 | GO:0005975 |
| d2f2ha1 | A | P31434 | GO:0016798 |
| d2f2ha1 | A | P31434 | GO:0030246 |
| d2f2ha1 | A | P31434 | GO:0042802 |
| d2f2ha1 | A | P31434 | GO:0061634 |
| d2f2ha1 | A | P31434 | GO:0080176 |
| d2f2ha1 | A | P31434 | GO:0003824 |
| d2f2ha1 | A | P31434 | GO:0004553 |
| d2f2ha1 | A | P31434 | GO:0005975 |
| d2f2ha1 | A | P31434 | GO:0016798 |
| d2f2ha1 | A | P31434 | GO:0030246 |
| d2f2ha1 | A | P31434 | GO:0042802 |
| d2f2ha1 | A | P31434 | GO:0061634 |
| d2f2ha1 | A | P31434 | GO:0080176 |
| d2f2ha1 | A | P31434 | GO:0003824 |
| d2f2ha1 | A | P31434 | GO:0004553 |
| d2f2ha1 | A | P31434 | GO:0005975 |
| d2f2ha1 | A | P31434 | GO:0016798 |
| d2f2ha1 | A | P31434 | GO:0030246 |
| d2f2ha1 | A | P31434 | GO:0042802 |
| d2f2ha1 | A | P31434 | GO:0061634 |
| d2f2ha1 | A | P31434 | GO:0080176 |
