solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1r1ma- ( 68 ) pqyvdetislsaktL---Fgfdkdslra
d2hqsc2 ( 68 ) nniVyFdldkydirs
d3oona1 ( 268 ) aIeveknnkGinLsFd-----IeFypnsfqIlq
d3td3a1 ( 221 ) eltedlnmeL----rVfFdtnksnikd
d4rhaa- ( 193 ) vqtkhftlkSdvL---Fnfnkstlkp
d4zhwa- ( 61 ) gmsskvlFgnnldrlnp
d5wtpa1 ( 335 ) ePdekeqkqLnqyAk--tIlFd--------------------tgkatikf
d6jfwa1 ( 1 ) d---kqeaeLrrqMegtgVeVqrqgddikLIMpGniT---Fatdsaniap
d7rjja- ( 428 ) pdvirLd----sslFdtgkwvlkp
60 70 80 90 100
d1r1ma- ( 93 ) eAqdnLkvlAqrLsrtnIqsVrVEGHtdfmgsdkyNqalSerrAyvVann
d2hqsc2 ( 83 ) dfaqmLdahAnfLrsnpsykVtVeGHADergtpeyNislGerRAnaVkmy
d3oona1 ( 296 ) keykkIdlIaklLekfkknnIlIeGHteqfgleee-helSekRAraIGny
d3td3a1 ( 244 ) qykpeIakVAekLseypnAtArIeGHTDntgprklNerlSlaRAnsVksa
d4rhaa- ( 216 ) eGqqaLdqLysqlsn--pgsvvVlGfTdrigsdayNqglSekrAqsVVdy
d4zhwa- ( 78 ) dsrntltkiArallavdidkvrleGHtdnygdegynqklSerRAesvAav
d5wtpa1 ( 363 ) qSaevLnqIinvLkkypnsrFrIEGHTdstgkkaknmilSqnrAdaVkvy
d6jfwa1 ( 45 ) sFyapLnnLAnsFkqynqntIeIVGYtdstgsrqhNmdlSqrrAqsVagy
d7rjja- ( 449 ) gStkrlvssl-dikarpgwlIvVaGHTDsvgeekaNqllSlkRAesvrdw
aaaaaaaaaaa bbbbb aaaaaaaaaaaaaaaaaa
110 120 130 140 150
d1r1ma- ( 143 ) Lvs-ngVpvsrIsavGlGesqaqmtqvÇeaevaklgakvskakkrealia
d2hqsc2 ( 133 ) Lqg-kgVsadqIsivSyGkekpavlgh--------------------dea
d3oona1 ( 346 ) Lik-kvKdkdqIlfkGwgsqk-----------------------------
d3td3a1 ( 294 ) LvneynvdasrLstqGfAwdqpiadnk--------------------tke
d4rhaa- ( 268 ) Lis-kgipsdkIsarG-gesnpvtgntÇdnvp------------raalid
d4zhwa- ( 128 ) Fre-AgMpaanievrGlGmskpvadnk--------------------tra
d5wtpa1 ( 413 ) Liq-ggIdagrLeSqGfGpekpiasnk--------------------nkk
d6jfwa1 ( 95 ) Lta-qgVdgtrLstrGmGpdqpiasns--------------------tad
d7rjja- ( 500 ) -rdtgdVpdsÇFavqGygesrpiatnd--------------------tpe
aaa 333bbbbb aa
160 170
d1r1ma- ( 192 ) çIepD-rrVDVkIrsiv
d2hqsc2 ( 162 ) AyskN-RrAvLvy
d3oona1 ( 377 ) ---aknrrVeITIln
d3td3a1 ( 324 ) gramN-rRVfAtItgsr
d4rhaa- ( 306 ) çLapD-rRVeIevkgvdvv
d4zhwa- ( 157 ) grsen-rRvaiivpa
d5wtpa1 ( 442 ) grelN-rrVeInli
d6jfwa1 ( 124 ) graqN-rRVEVnLrpv---p
d7rjja- ( 529 ) gralN-rRVeIsLvpqvdaç
a bbbbbbb
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| Outer membrane protein class 4, RmpM, C-terminal domain | OmpA-like | Neisseria meningitidis [TaxId: 487] | 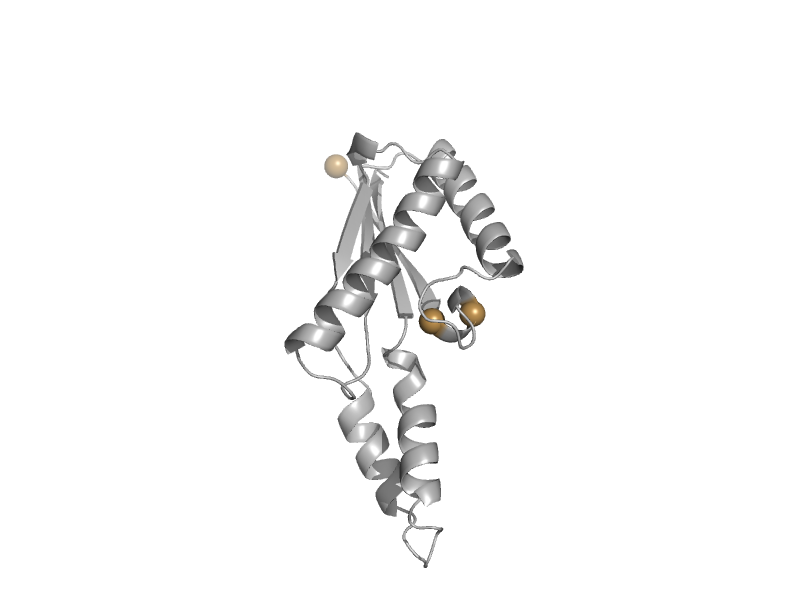 | view | |
| automated matches | OmpA-like | Escherichia coli [TaxId: 562] | 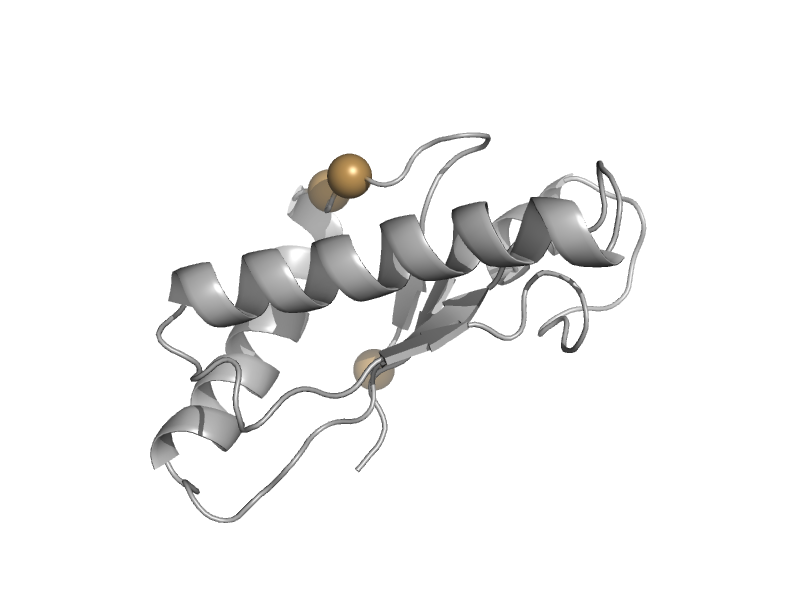 | view | |
| automated matches | automated matches | Borrelia burgdorferi [TaxId: 224326] | 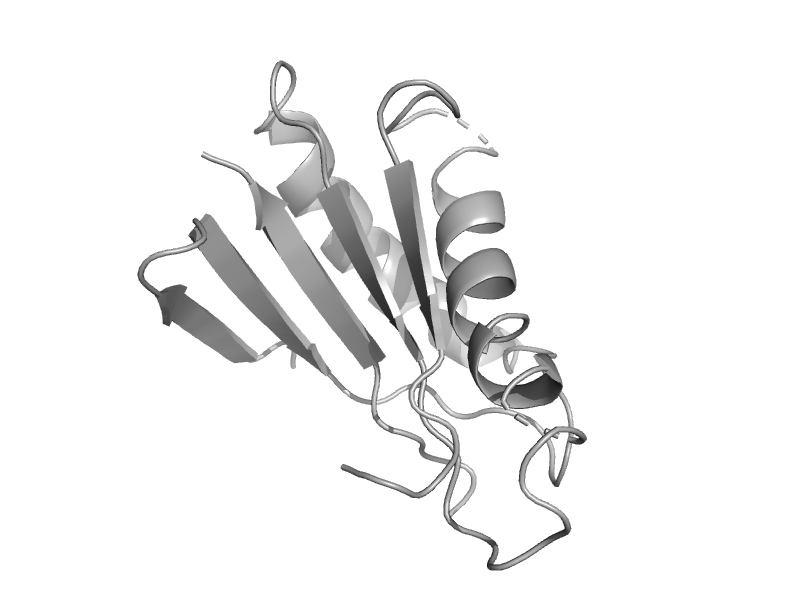 | view | |
| automated matches | automated matches | Acinetobacter baumannii [TaxId: 470] | 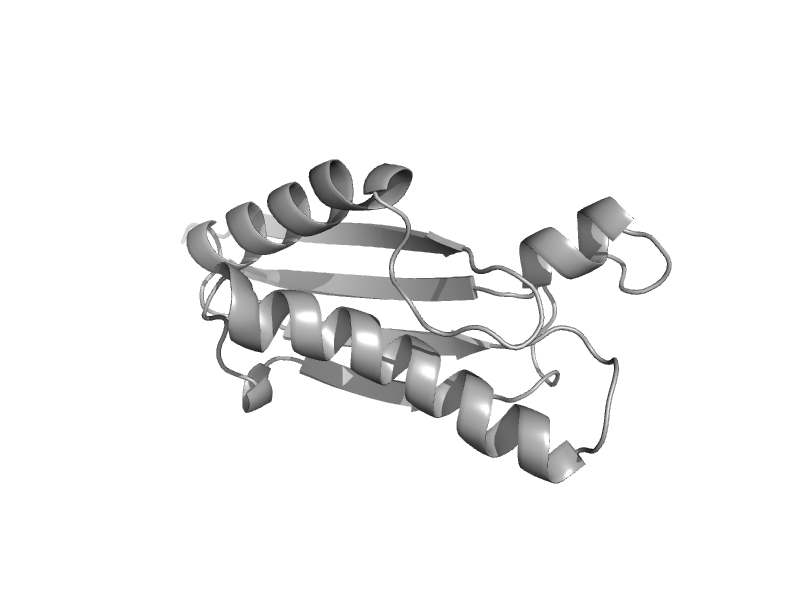 | view | |
| automated matches | automated matches | Salmonella enterica [TaxId: 99287] |  | view | |
| automated matches | automated matches | Pseudomonas aeruginosa [TaxId: 208964] |  | view | |
| automated matches | automated matches | Capnocytophaga gingivalis [TaxId: 553178] |  | view | |
| automated matches | automated matches | Pseudomonas aeruginosa [TaxId: 208964] |  | view | |
| automated matches | automated matches | Klebsiella pneumoniae [TaxId: 72407] |  | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | 150 | 160 | 170 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | P | Q | Y | V | D | E | T | I | S | L | S | A | K | T | L | - | - | - | F | G | F | D | K | D | S | L | R | A | E | A | Q | D | N | L | K | V | L | A | Q | R | L | S | R | T | N | I | Q | S | V | R | V | E | G | H | T | D | F | M | G | S | D | K | Y | N | Q | A | L | S | E | R | R | A | Y | V | V | A | N | N | L | V | S | - | N | G | V | P | V | S | R | I | S | A | V | G | L | G | E | S | Q | A | Q | M | T | Q | V | C | E | A | E | V | A | K | L | G | A | K | V | S | K | A | K | K | R | E | A | L | I | A | C | I | E | P | D | - | R | R | V | D | V | K | I | R | S | I | V | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | N | N | I | V | Y | F | D | L | D | K | Y | D | I | R | S | D | F | A | Q | M | L | D | A | H | A | N | F | L | R | S | N | P | S | Y | K | V | T | V | E | G | H | A | D | E | R | G | T | P | E | Y | N | I | S | L | G | E | R | R | A | N | A | V | K | M | Y | L | Q | G | - | K | G | V | S | A | D | Q | I | S | I | V | S | Y | G | K | E | K | P | A | V | L | G | H | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | D | E | A | A | Y | S | K | N | - | R | R | A | V | L | V | Y | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | A | I | E | V | E | K | N | N | K | G | I | N | L | S | F | D | - | - | - | - | - | I | E | F | Y | P | N | S | F | Q | I | L | Q | K | E | Y | K | K | I | D | L | I | A | K | L | L | E | K | F | K | K | N | N | I | L | I | E | G | H | T | E | Q | F | G | L | E | E | E | - | H | E | L | S | E | K | R | A | R | A | I | G | N | Y | L | I | K | - | K | V | K | D | K | D | Q | I | L | F | K | G | W | G | S | Q | K | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | A | K | N | R | R | V | E | I | T | I | L | N | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | E | L | T | E | D | L | N | M | E | L | - | - | - | - | R | V | F | F | D | T | N | K | S | N | I | K | D | Q | Y | K | P | E | I | A | K | V | A | E | K | L | S | E | Y | P | N | A | T | A | R | I | E | G | H | T | D | N | T | G | P | R | K | L | N | E | R | L | S | L | A | R | A | N | S | V | K | S | A | L | V | N | E | Y | N | V | D | A | S | R | L | S | T | Q | G | F | A | W | D | Q | P | I | A | D | N | K | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | T | K | E | G | R | A | M | N | - | R | R | V | F | A | T | I | T | G | S | R | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | V | Q | T | K | H | F | T | L | K | S | D | V | L | - | - | - | F | N | F | N | K | S | T | L | K | P | E | G | Q | Q | A | L | D | Q | L | Y | S | Q | L | S | N | - | - | P | G | S | V | V | V | L | G | F | T | D | R | I | G | S | D | A | Y | N | Q | G | L | S | E | K | R | A | Q | S | V | V | D | Y | L | I | S | - | K | G | I | P | S | D | K | I | S | A | R | G | - | G | E | S | N | P | V | T | G | N | T | C | D | N | V | P | - | - | - | - | - | - | - | - | - | - | - | - | R | A | A | L | I | D | C | L | A | P | D | - | R | R | V | E | I | E | V | K | G | V | D | V | V | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | M | S | S | K | V | L | F | G | N | N | L | D | R | L | N | P | D | S | R | N | T | L | T | K | I | A | R | A | L | L | A | V | D | I | D | K | V | R | L | E | G | H | T | D | N | Y | G | D | E | G | Y | N | Q | K | L | S | E | R | R | A | E | S | V | A | A | V | F | R | E | - | A | G | M | P | A | A | N | I | E | V | R | G | L | G | M | S | K | P | V | A | D | N | K | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | T | R | A | G | R | S | E | N | - | R | R | V | A | I | I | V | P | A | - | - | - | - | - |
| E | P | D | E | K | E | Q | K | Q | L | N | Q | Y | A | K | - | - | T | I | L | F | D | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | T | G | K | A | T | I | K | F | Q | S | A | E | V | L | N | Q | I | I | N | V | L | K | K | Y | P | N | S | R | F | R | I | E | G | H | T | D | S | T | G | K | K | A | K | N | M | I | L | S | Q | N | R | A | D | A | V | K | V | Y | L | I | Q | - | G | G | I | D | A | G | R | L | E | S | Q | G | F | G | P | E | K | P | I | A | S | N | K | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | N | K | K | G | R | E | L | N | - | R | R | V | E | I | N | L | I | - | - | - | - | - | - |
| D | - | - | - | K | Q | E | A | E | L | R | R | Q | M | E | G | T | G | V | E | V | Q | R | Q | G | D | D | I | K | L | I | M | P | G | N | I | T | - | - | - | F | A | T | D | S | A | N | I | A | P | S | F | Y | A | P | L | N | N | L | A | N | S | F | K | Q | Y | N | Q | N | T | I | E | I | V | G | Y | T | D | S | T | G | S | R | Q | H | N | M | D | L | S | Q | R | R | A | Q | S | V | A | G | Y | L | T | A | - | Q | G | V | D | G | T | R | L | S | T | R | G | M | G | P | D | Q | P | I | A | S | N | S | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | T | A | D | G | R | A | Q | N | - | R | R | V | E | V | N | L | R | P | V | - | - | - | P |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | P | D | V | I | R | L | D | - | - | - | - | S | S | L | F | D | T | G | K | W | V | L | K | P | G | S | T | K | R | L | V | S | S | L | - | D | I | K | A | R | P | G | W | L | I | V | V | A | G | H | T | D | S | V | G | E | E | K | A | N | Q | L | L | S | L | K | R | A | E | S | V | R | D | W | - | R | D | T | G | D | V | P | D | S | C | F | A | V | Q | G | Y | G | E | S | R | P | I | A | T | N | D | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | T | P | E | G | R | A | L | N | - | R | R | V | E | I | S | L | V | P | Q | V | D | A | C |
| Member | Chain | UniProtKB | GO term |
|---|---|---|---|
| d1r1ma_ | A | P0A0V3 | GO:0009279 |
| d1r1ma_ | A | P0A0V3 | GO:0016020 |
