solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1olta- ( 4 ) qqIdwdlalikyytsYPtaleFsedfgeqaFlqAvary--perpLSLyVh
d1r30a- ( 5 ) p--rwtlsqVtelF------ekplldLlfeAqqvhrqhfdprqVqvStll
d1tv8a- ( 3 ) eqikDklgRpIrdLr
bbb
60 70 80 90 100
d1olta- ( 59 ) IPFc---hklcyfcgcnkivtr----qqhkAdqYLdaLeqEIvhrAplFa
d1r30a- ( 47 ) SIktGacpedckycpqSsrytg----leaerlmeveqVlesArkakaa--
d1tv8a- ( 18 ) LsVTdrcnFrcdycmpkevfgddfvflpkneLLtfdeMarIAkVYAeL--
b aaaaaaaaaaa
110 120 130 140 150
d1olta- ( 102 ) gRhVsqLhWGGGtPTYL-nkaQIsrLMklLrenFqFnadAeIsIeVdPre
d1r30a- ( 92 ) --gStRFcMGAawnPherdMpyLeqMVqgVkam-----gLeACMtLgt--
d1tv8a- ( 66 ) --gVkkIrItGGEPLmrr---dLdvLIakLnqid----gIedIGLtTnGl
bbbbb aaaaaaaa bbbb
160 170 180 190 200
d1olta- ( 151 ) IeldvLdhLraegFnrLsMgVqDFnkeVQrl-VnreQdeefIfaLLnhAr
d1r30a- ( 134 ) LsesqAqrLanagLdYYnHnLDT-SpefYgn-IIttrtyqerldTLekVr
d1tv8a- ( 107 ) lLkkhGqkLydagLrrInVsLdAiddtlFqsiNnrnikAttIleqIdyAt
aaaaaaaa bbb aaaaaa a aaaaaaaaaaaa
210 220 230 240 250
d1olta- ( 200 ) eigFtsTnIdLiyGLPkQtpesFafTLkrVaeL--nPdrLsVfnYahlpt
d1r30a- ( 182 ) dAgI-kVCSGGiVgL-gEtvkdrAglLlqLAnlptpPeSVPInmLvkvk-
d1tv8a- ( 157 ) sigL-nVkVnVvIqk-giNddqIipMLeyFkd--khieIrFiEfmdvgn-
a b bbb aaaaaaaaa bbbbb
260 270 280 290 300
d1olta- ( 248 ) ifaaQrkIkdadlPspqqkldIlqeTIafLtqs--------gYqFIGMdh
d1r30a- ( 229 ) -gTpladnd---dvdafdFIrTIAVARiMMp----------tSyVRLS--
d1tv8a- ( 202 ) ----------dnGWd-fskVVtkdeMltmIeqhfeIdpvepkyfgEvAkY
aaaaaaa
310 320 330 340 350
d1olta- ( 290 ) FA-rpd--deLAvaqregvLhrnf---qgYttqgdtDlLGMGVsAiSmIG
d1r30a- ( 263 ) aG-ReqMneqtQamCfmAGANSIf-----Ygc--------kllttpNPe-
d1tv8a- ( 241 ) yrHk-----------dngvqFGLiTsVsqSfcstctrArLssdGkfyGcl
b
360 370 380 390 400
d1olta- ( 334 ) dcYAqnqkelkqYyqqVdeqGnALwrGiaLtrdDcIRrdVIksLICnFrL
d1r30a- ( 298 ) ---------edkDlqLFrklglnpqqt
d1tv8a- ( 280 ) fatvdgfnVkafIrs----------------gvtdeeLkeqFkalWqiRd
aaaaa
410 420 430 440 450
d1olta- ( 384 ) dYspieqqwdllFadyFaeDlklLaplakdgLVdvdekgIqVtakGrllI
d1r30a-
d1tv8a- ( 314 ) ---------drysdertaqtvanrq
460
d1olta- ( 434 ) rnICmcFDtyl
d1r30a-
d1tv8a-
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| Oxygen-independent coproporphyrinogen III oxidase HemN | Oxygen-independent coproporphyrinogen III oxidase HemN | Escherichia coli [TaxId: 562] | 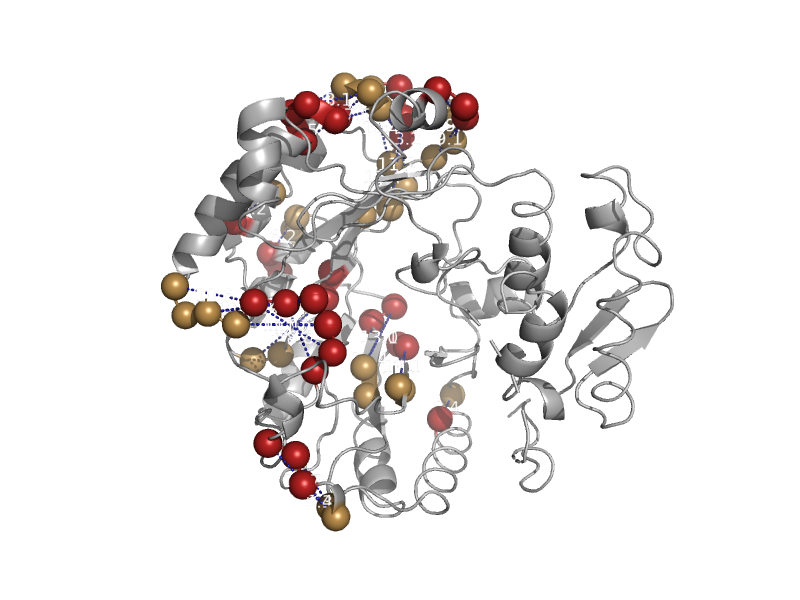 | view | |
| Biotin synthase | Biotin synthase | Escherichia coli [TaxId: 562] | 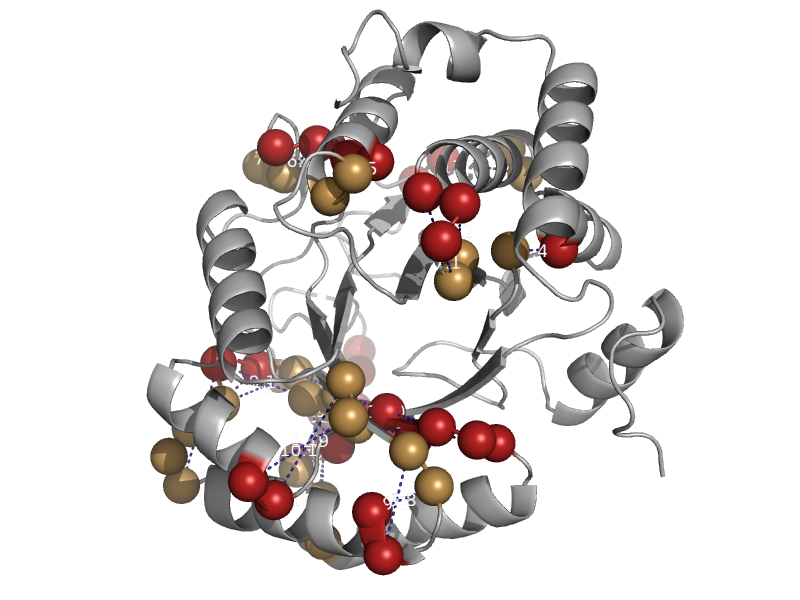 | view | |
| Molybdenum cofactor biosynthesis protein A MoaA | MoCo biosynthesis proteins | Staphylococcus aureus [TaxId: 1280] | 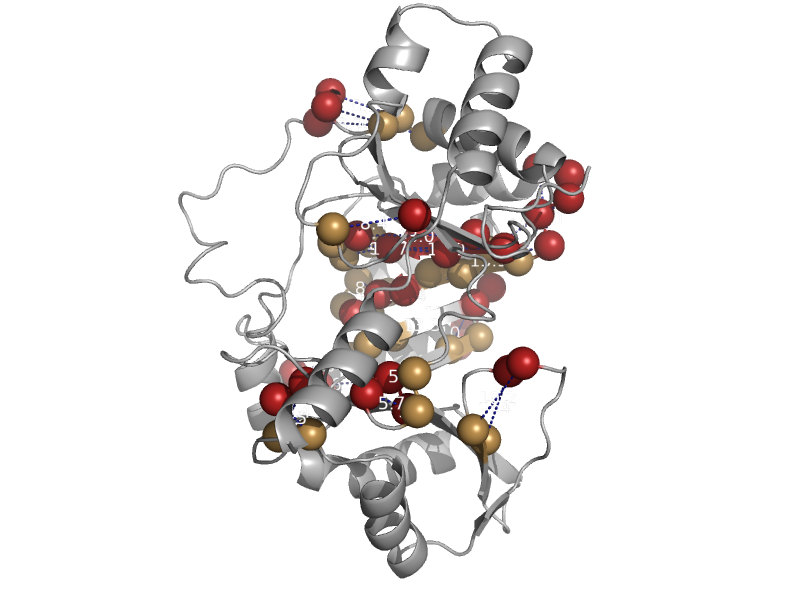 | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | 100 | 110 | 120 | 130 | 140 | 150 | 160 | 170 | 180 | 190 | 200 | 210 | 220 | 230 | 240 | 250 | 260 | 270 | 280 | 290 | 300 | 310 | 320 | 330 | 340 | 350 | 360 | 370 | 380 | 390 | 400 | 410 | 420 | 430 | 440 | 450 | 460 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Q | Q | I | D | W | D | L | A | L | I | K | Y | Y | T | S | Y | P | T | A | L | E | F | S | E | D | F | G | E | Q | A | F | L | Q | A | V | A | R | Y | - | - | P | E | R | P | L | S | L | Y | V | H | I | P | F | C | - | - | - | H | K | L | C | Y | F | C | G | C | N | K | I | V | T | R | - | - | - | - | Q | Q | H | K | A | D | Q | Y | L | D | A | L | E | Q | E | I | V | H | R | A | P | L | F | A | G | R | H | V | S | Q | L | H | W | G | G | G | T | P | T | Y | L | - | N | K | A | Q | I | S | R | L | M | K | L | L | R | E | N | F | Q | F | N | A | D | A | E | I | S | I | E | V | D | P | R | E | I | E | L | D | V | L | D | H | L | R | A | E | G | F | N | R | L | S | M | G | V | Q | D | F | N | K | E | V | Q | R | L | - | V | N | R | E | Q | D | E | E | F | I | F | A | L | L | N | H | A | R | E | I | G | F | T | S | T | N | I | D | L | I | Y | G | L | P | K | Q | T | P | E | S | F | A | F | T | L | K | R | V | A | E | L | - | - | N | P | D | R | L | S | V | F | N | Y | A | H | L | P | T | I | F | A | A | Q | R | K | I | K | D | A | D | L | P | S | P | Q | Q | K | L | D | I | L | Q | E | T | I | A | F | L | T | Q | S | - | - | - | - | - | - | - | - | G | Y | Q | F | I | G | M | D | H | F | A | - | R | P | D | - | - | D | E | L | A | V | A | Q | R | E | G | V | L | H | R | N | F | - | - | - | Q | G | Y | T | T | Q | G | D | T | D | L | L | G | M | G | V | S | A | I | S | M | I | G | D | C | Y | A | Q | N | Q | K | E | L | K | Q | Y | Y | Q | Q | V | D | E | Q | G | N | A | L | W | R | G | I | A | L | T | R | D | D | C | I | R | R | D | V | I | K | S | L | I | C | N | F | R | L | D | Y | S | P | I | E | Q | Q | W | D | L | L | F | A | D | Y | F | A | E | D | L | K | L | L | A | P | L | A | K | D | G | L | V | D | V | D | E | K | G | I | Q | V | T | A | K | G | R | L | L | I | R | N | I | C | M | C | F | D | T | Y | L |
| P | - | - | R | W | T | L | S | Q | V | T | E | L | F | - | - | - | - | - | - | E | K | P | L | L | D | L | L | F | E | A | Q | Q | V | H | R | Q | H | F | D | P | R | Q | V | Q | V | S | T | L | L | S | I | K | T | G | A | C | P | E | D | C | K | Y | C | P | Q | S | S | R | Y | T | G | - | - | - | - | L | E | A | E | R | L | M | E | V | E | Q | V | L | E | S | A | R | K | A | K | A | A | - | - | - | - | G | S | T | R | F | C | M | G | A | A | W | N | P | H | E | R | D | M | P | Y | L | E | Q | M | V | Q | G | V | K | A | M | - | - | - | - | - | G | L | E | A | C | M | T | L | G | T | - | - | L | S | E | S | Q | A | Q | R | L | A | N | A | G | L | D | Y | Y | N | H | N | L | D | T | - | S | P | E | F | Y | G | N | - | I | I | T | T | R | T | Y | Q | E | R | L | D | T | L | E | K | V | R | D | A | G | I | - | K | V | C | S | G | G | I | V | G | L | - | G | E | T | V | K | D | R | A | G | L | L | L | Q | L | A | N | L | P | T | P | P | E | S | V | P | I | N | M | L | V | K | V | K | - | - | G | T | P | L | A | D | N | D | - | - | - | D | V | D | A | F | D | F | I | R | T | I | A | V | A | R | I | M | M | P | - | - | - | - | - | - | - | - | - | - | T | S | Y | V | R | L | S | - | - | A | G | - | R | E | Q | M | N | E | Q | T | Q | A | M | C | F | M | A | G | A | N | S | I | F | - | - | - | - | - | Y | G | C | - | - | - | - | - | - | - | - | K | L | L | T | T | P | N | P | E | - | - | - | - | - | - | - | - | - | - | E | D | K | D | L | Q | L | F | R | K | L | G | L | N | P | Q | Q | T | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | E | Q | I | K | D | K | L | G | R | P | I | R | D | L | R | L | S | V | T | D | R | C | N | F | R | C | D | Y | C | M | P | K | E | V | F | G | D | D | F | V | F | L | P | K | N | E | L | L | T | F | D | E | M | A | R | I | A | K | V | Y | A | E | L | - | - | - | - | G | V | K | K | I | R | I | T | G | G | E | P | L | M | R | R | - | - | - | D | L | D | V | L | I | A | K | L | N | Q | I | D | - | - | - | - | G | I | E | D | I | G | L | T | T | N | G | L | L | L | K | K | H | G | Q | K | L | Y | D | A | G | L | R | R | I | N | V | S | L | D | A | I | D | D | T | L | F | Q | S | I | N | N | R | N | I | K | A | T | T | I | L | E | Q | I | D | Y | A | T | S | I | G | L | - | N | V | K | V | N | V | V | I | Q | K | - | G | I | N | D | D | Q | I | I | P | M | L | E | Y | F | K | D | - | - | K | H | I | E | I | R | F | I | E | F | M | D | V | G | N | - | - | - | - | - | - | - | - | - | - | - | D | N | G | W | D | - | F | S | K | V | V | T | K | D | E | M | L | T | M | I | E | Q | H | F | E | I | D | P | V | E | P | K | Y | F | G | E | V | A | K | Y | Y | R | H | K | - | - | - | - | - | - | - | - | - | - | - | D | N | G | V | Q | F | G | L | I | T | S | V | S | Q | S | F | C | S | T | C | T | R | A | R | L | S | S | D | G | K | F | Y | G | C | L | F | A | T | V | D | G | F | N | V | K | A | F | I | R | S | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | G | V | T | D | E | E | L | K | E | Q | F | K | A | L | W | Q | I | R | D | - | - | - | - | - | - | - | - | - | D | R | Y | S | D | E | R | T | A | Q | T | V | A | N | R | Q | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
