solvent inaccessible: UPPER CASE X
solvent accesible: lower case x
alpha helix: red x
solvent accesible: lower case x
alpha helix: red x
beta strand: blue x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
3 - 10 helix: maroon x
hydrogen bond to main chain amide: bold x
hydrogen bond to mainchain carbonyl: underline x
disulphide bond: cedilla
positive phi: italic x
disulphide bond: cedilla
positive phi: italic x
10 20 30 40 50
d1pj5a1 ( 743 ) arrlrCLtIddgrsiVlgkepVfyk---e-qavgyVtsAaygytvakPiA
d1v5va1 ( 313 ) rklvhFkMid-kgiPregykVyAn---g-emIgeVtsGtlSpllnvGig
d1vloa1 ( 278 ) teklVGLvte--kgvLrnelpVrftdaqgnqhegiItsGtfSptlgysiA
d1vlya1 ( 244 ) nralwlLagsA-srlPeagedLelkg---nwrtgtVlaAvkled-gqVvV
d1wosa1 ( 279 ) rklVALeLs-gkriArkgyeVlkn---g-ervgeItsGnfsptlgkSiA
bbbbbbb bbbb bbbbbbbbbbb bbb
60 70 80 90
d1pj5a1 ( 789 ) ySyLpgtV-svgdsVdIeyf-grritAtVtedplydpkmtrl---g
d1v5va1 ( 357 ) iAfVkeeyAkpgieIeVeIr-gqrkkAvTvtppfydpkkyglfret
d1vloa1 ( 327 ) lArVpgIg----etAiVqirn---epvkvtkpvfvr----ngkava
d1vlya1 ( 293 ) qvvnnd---epdsifrVr---dDantLhieplp-ysl
d1wosa1 ( 323 ) lAlVsksV-kigdqLgVvFpggklveAlVvkkpfy---rgsv---r
bbbb bbb bbbb
| Domain ID | Name | Family | Source | Domain | STRING-DB |
|---|---|---|---|---|---|
| N,N-dimethylglycine oxidase, C-terminal domain | Aminomethyltransferase beta-barrel domain | Arthrobacter globiformis [TaxId: 1665] | 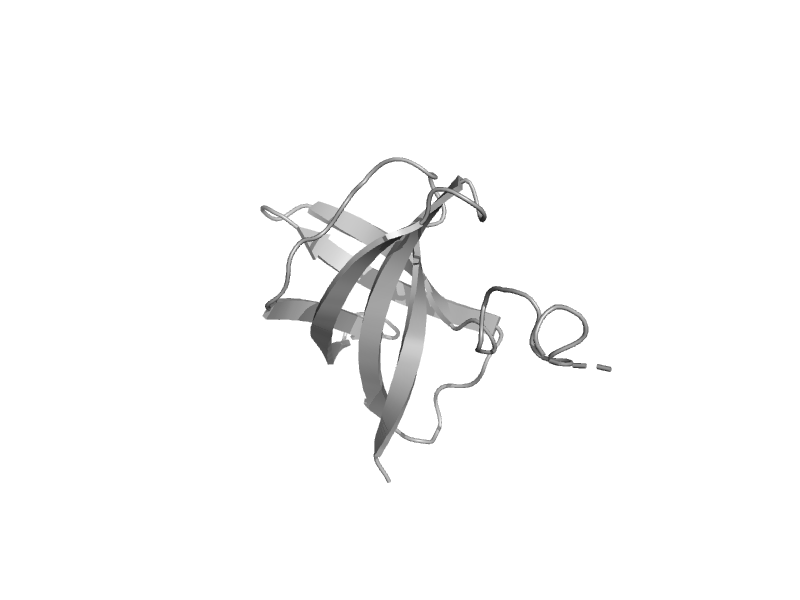 | view | |
| Glycine cleavage system T protein, GcvT | Aminomethyltransferase beta-barrel domain | Pyrococcus horikoshii [TaxId: 53953] |  | view | |
| Glycine cleavage system T protein, GcvT | Aminomethyltransferase beta-barrel domain | Escherichia coli [TaxId: 562] | 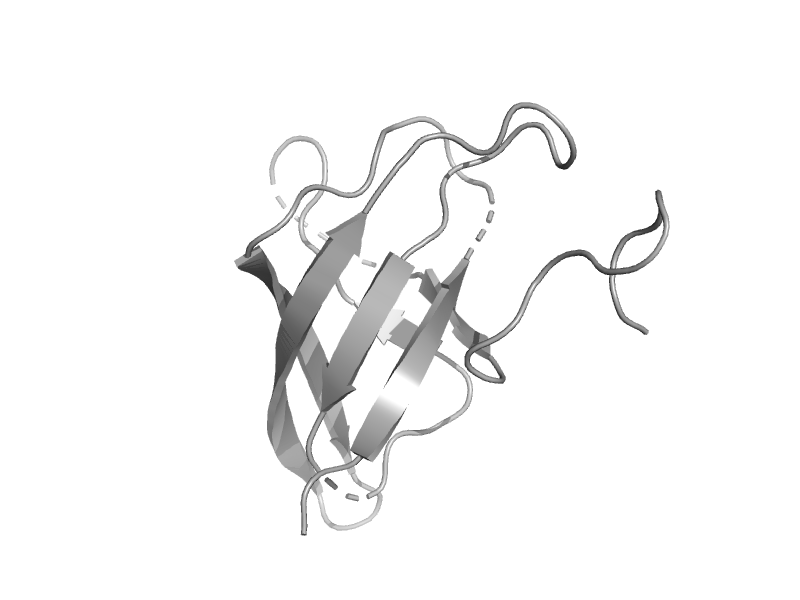 | view | |
| Hypothetical protein YgfZ, C-terminal domain | Aminomethyltransferase beta-barrel domain | Escherichia coli [TaxId: 562] | 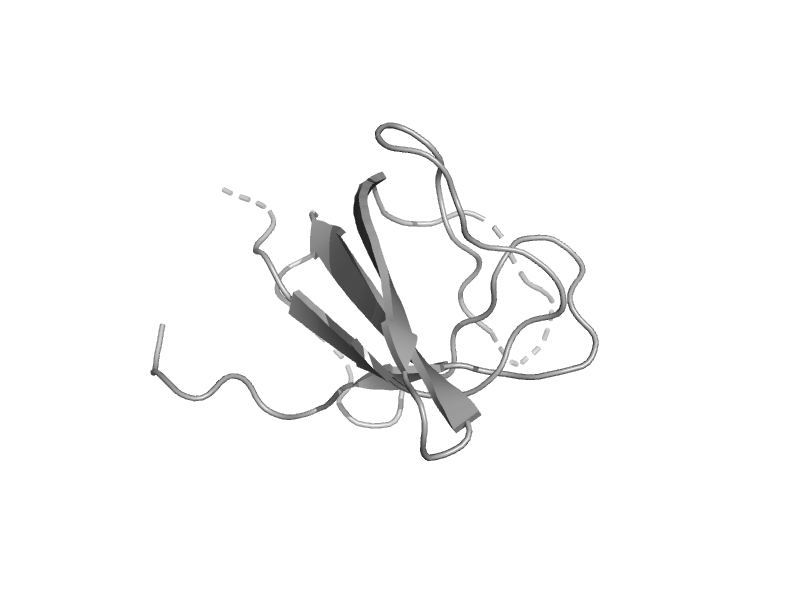 | view | |
| Glycine cleavage system T protein, GcvT | Aminomethyltransferase beta-barrel domain | Thermotoga maritima [TaxId: 2336] | 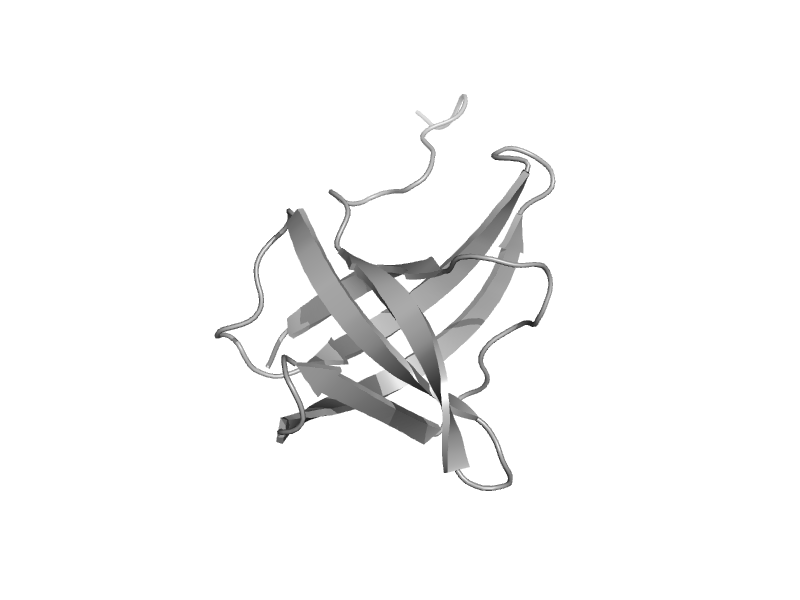 | view |
No outliers
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 80 | 90 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | R | R | L | R | C | L | T | I | D | D | G | R | S | I | V | L | G | K | E | P | V | F | Y | K | - | - | - | E | - | Q | A | V | G | Y | V | T | S | A | A | Y | G | Y | T | V | A | K | P | I | A | Y | S | Y | L | P | G | T | V | - | S | V | G | D | S | V | D | I | E | Y | F | - | G | R | R | I | T | A | T | V | T | E | D | P | L | Y | D | P | K | M | T | R | L | - | - | - | G |
| - | R | K | L | V | H | F | K | M | I | D | - | K | G | I | P | R | E | G | Y | K | V | Y | A | N | - | - | - | G | - | E | M | I | G | E | V | T | S | G | T | L | S | P | L | L | N | V | G | I | G | I | A | F | V | K | E | E | Y | A | K | P | G | I | E | I | E | V | E | I | R | - | G | Q | R | K | K | A | V | T | V | T | P | P | F | Y | D | P | K | K | Y | G | L | F | R | E | T |
| T | E | K | L | V | G | L | V | T | E | - | - | K | G | V | L | R | N | E | L | P | V | R | F | T | D | A | Q | G | N | Q | H | E | G | I | I | T | S | G | T | F | S | P | T | L | G | Y | S | I | A | L | A | R | V | P | G | I | G | - | - | - | - | E | T | A | I | V | Q | I | R | N | - | - | - | E | P | V | K | V | T | K | P | V | F | V | R | - | - | - | - | N | G | K | A | V | A |
| N | R | A | L | W | L | L | A | G | S | A | - | S | R | L | P | E | A | G | E | D | L | E | L | K | G | - | - | - | N | W | R | T | G | T | V | L | A | A | V | K | L | E | D | - | G | Q | V | V | V | Q | V | V | N | N | D | - | - | - | E | P | D | S | I | F | R | V | R | - | - | - | D | D | A | N | T | L | H | I | E | P | L | P | - | Y | S | L | - | - | - | - | - | - | - | - | - |
| - | R | K | L | V | A | L | E | L | S | - | G | K | R | I | A | R | K | G | Y | E | V | L | K | N | - | - | - | G | - | E | R | V | G | E | I | T | S | G | N | F | S | P | T | L | G | K | S | I | A | L | A | L | V | S | K | S | V | - | K | I | G | D | Q | L | G | V | V | F | P | G | G | K | L | V | E | A | L | V | V | K | K | P | F | Y | - | - | - | R | G | S | V | - | - | - | R |
