RNA-binding proteins (RBPs) stabilise, protect, package, transport and mediate interactions with or act catalytically on RNA (cutting, unwinding, replicating, modifying, etc.) [1]. The association of this class of proteins with their RNA partners is dynamic and defines the cellular localisation, lifetime and processing of different kinds of RNA, as well as the rate of translation of mRNA [2]. EcRBPome is a comprehensive database that documents Escherichia coli RBPs. All the data presented in this database were obtained from our previous study [3], in which genome-wide survey (GWS) of RBPs were performed for 166 complete E. coli proteomes, retrieved from the RefSeq database (May 2016). The start-points for such search methods, were known sequence and structure signatures of RBPs, organised as structure-centric and sequence-centric family Hidden Markov Models (HMMs) [4]. The structure-centric RBP families were obtained using structure-based sequence alignments of known RBP structures, deposited in the PDB in complex with RNA. The sequence-centric RBP families were retrieved from the Pfam 28 database, on the basis of keyword search.
|
|
|
|
|
|
|
|
|
|
|
- Browse
- Browse all strains in EcRBPome: List of all the E. coli strains present in this database, with links to the assembly, biosample and bioproject details for each strain. The pathogenicity is recorded along with the pathotype information (AIEC: Adherent Invasive E. coli, APEC: Avian Pathogenic E. coli, EAEC: EnteroAggregative E. coli, EHEC: EnteroHemorrhagic E. coli, EIEC: EnteroInvasive E. coli, EPEC: EnteroPathogenic E. coli, ETEC: EnteroToxigenic E. coli, ExPEC: Extraintestinal Pathogenic E. coli, NMEC: Neonatal Meningitis E. coli, STEC: Shiga Toxin producing E. coli, UPEC: UroPathogenic E. coli). The sequence type is derived from the MLST information available at the Enterobase[5].
- Browse all RBPs in EcRBPome: List of all the E. coli RNA-binding proteins (RBPs) present in this database, with links to the RefSeq page and downloadable FASTA sequence for each RBP. The sequences of the RBPs can also be submitted directly to RStrucFam [6], for the prediction of their function and cognate RNA partner(s).
- Browse all RBP domain architectures in EcRBPome: Provides the lists the Pfam28 domain architectures (DAs) for all the RBPs present in this database, the distribution of single RBDs
and the distribution of pairs of domains architectures as separate tabs. Pathogen specific RBDs and pathogen specific domain architectures have been highlighted in red and nonpathogen specific RBDs,
nonpathogen specific domain architectures have been highlighted in green, RBDs have been highlighted in bold also and the asterisk (*) is showing the interesting domain architectures or RBDs that have been choosen because they are showing the difference between the three numbers
(Number of occurrences in proteins present in both pathogenic and nonpathogenic strains, Number of occurrences in pathogen-specific proteins, Number of occurrences in nonpathogen-specific proteins). All the DAs have been highlighted in black font and the RNA-binding domains (RBDs) in bold also.
In the "All RBP domain architecture" tab, it is to be noted that in Pfamscan for identification of domains, a few of the RBPs (for eg. NP_052625.1) associate with all non-RBDs in their DA.
These RBP DAs have been highlighted with an asterisk (*). In such cases, non-RBDs associate to the protein sequence with higher confidence (on the basis E-value and domain scores), as compared to RBDs in overlapping regions, when queried with HMMs from the entire Pfam, but associate with RBDs with acceptable E-value and domain scores when queried with Pfam RBD HMMs alone. These RBPs are not regarded as false positives, since many of them have RNA-binding evidence. It is also possible that the non-RBDs are not annotated with any RNA-binding property yet, but may be associated with such function in future.
- Cross-strain comparisons
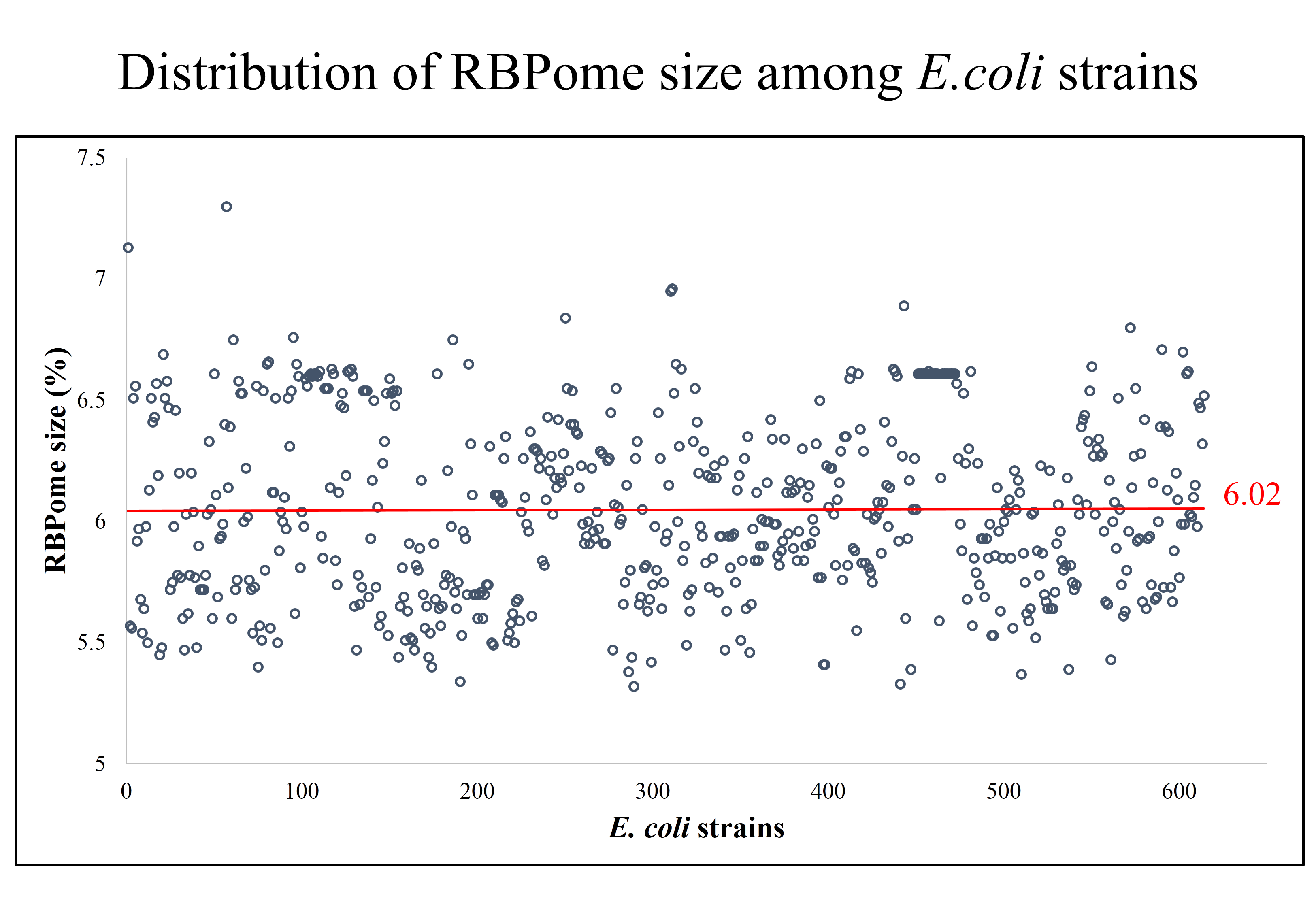
- Percentage of RBPs: Lists the percentage of RBPs in each E. coli proteome, with links to the details of each strain. It also provides comparative graphical representations of the percentage of RBPs in each strain versus the average of that across all strains, available for download by the user.
- Cross-strain RBD distribution: Depicts the presence or absence of all RBDs across all E. coli strains in the form of a matrix.
- Strain-centric RBD distribution: Depicts the RBD composition of each E. coli strain in a graphical manner. The percentage of the various RBDs in each of the strains are also provided as a downloadable flatfile.
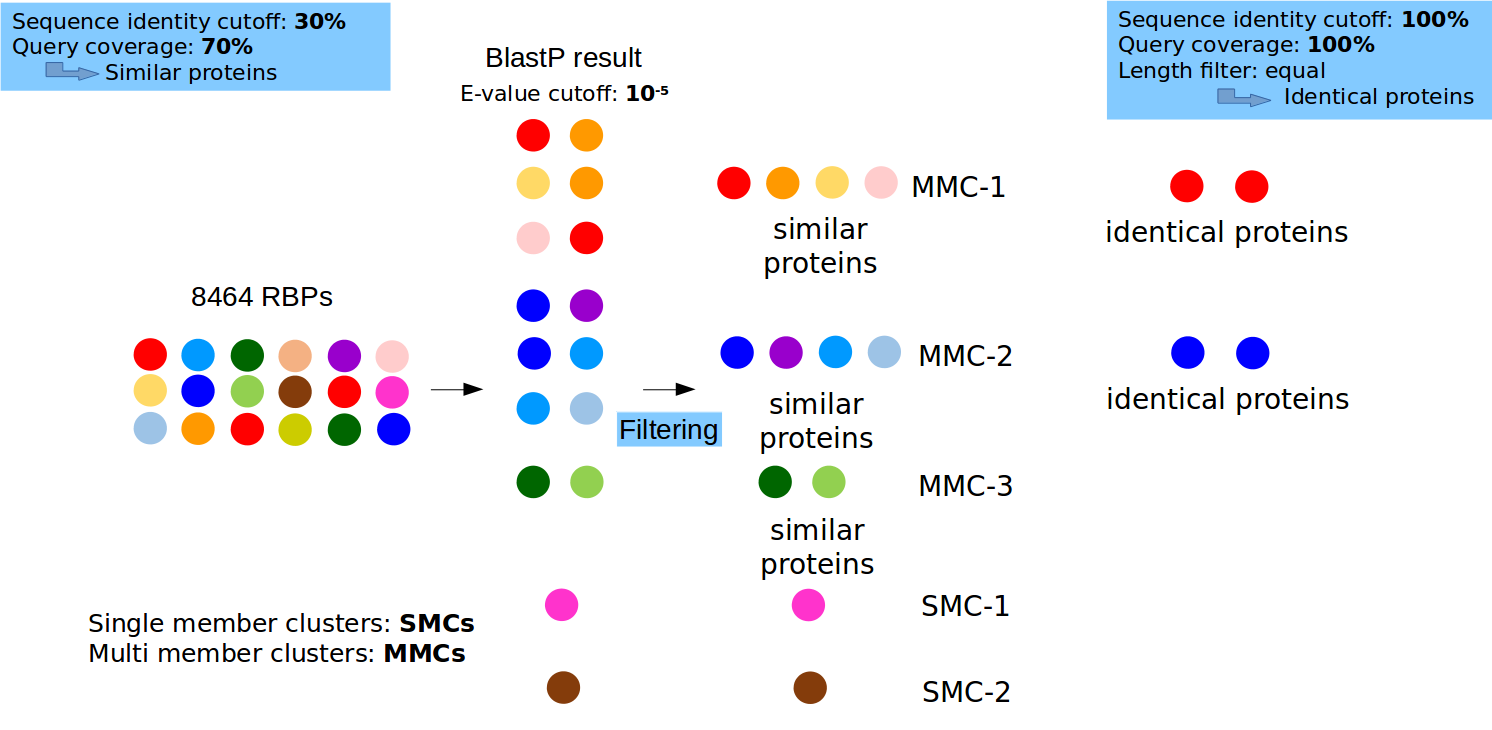
- Single-link clustering of all RBPs: Provides the results of single-link clustering of identical and similar proteins as separate tabs. In each table, details about the proteins involved, the information about their presence in pathogen-specific or nonpathogen-specific strains, a link to download the PSI-BLAST results, if any (E-value cutoff = 10-5, H-value cutoff = 10-5, number of iterations = 5) for each cluster is provided.
- Cross-reference to other databases
- Links to UniProt and PDB: Provides mappings to the UniProt and the PDB for each RBP (RefSeq ID).
- GO annotations: Lists the available Gene ontology (GO) annotations (biological processes, molecular functions and cellular componenents) for all the RBPs in this database.
- EC annotations: Lists the available Enzyme Commission (EC) numbers for all the RBPs in this database and link to the database BRENDA.
- Download sequences
- Download all RBP sequences: Link to download the FASTA sequences of all the RBPs present in this database.
- Download RBP sequences for each strain: Link to download the FASTA sequences of all the RBPs encoded in each strain present in this database.
- Download all RBD sequences: Link to download the FASTA sequences of all the RBDs predicted to be encoded in the RBPs present in this database.
- Download all RBP DAs: Link to download all the Pfam28 domain architectures (DAs) for all the RBPs present in this database.
- Download all files: Link to download all the files as one compressed '.tar.gz' file.
- CAPS: Link to the lab webpage of Prof. R. Sowdhamini at National Centre for Biological Sciences (NCBS).
- RStrucFam: Link to the in-house RStrucFam webserver, to associate structure and cognate RNA for RBPs from sequence information.[6]
- Feedback: This page allows the users to give suggestions about EcRBPome. Please do not hesitate to write back to us! :-)
[1] Dreyfuss, G., Kim, V.N. and Kataoka, N. (2002) Messenger-RNA-binding proteins and the
messages they carry. Nature reviews. Molecular cell biology, 3, 195-205.
[2] Lunde, B.M., Moore, C. and Varani, G. (2007) RNA-binding proteins: modular design for efficient function. Nature reviews. Molecular cell biology, 8, 479-490.
[3] Ghosh, P., and Sowdhamini, R. (2017) Bioinformatics comparisons of RNA-binding proteins of pathogenic and non-pathogenic Escherichia coli strains reveal novel virulence factors. BMC Genomics, 18(1), 658.
[4] Ghosh, P. and Sowdhamini, R. (2016) Genome-wide survey of putative RNA-binding
proteins encoded in the human proteome. Mol. BioSyst., 12, 532-540.
[5] Alikhan, N.F., Zhou, Z., Sergeant, M.J. and Achtman, M. (2018) A genomic overview
of the population structure of Salmonella. PLoS Genet., 14(4), e1007261.
[6] Ghosh, P., Mathew, O.K. and Sowdhamini, R. (2016) RStrucFam: a web server to associate
structure and cognate RNA for RNA-binding proteins from sequence information. BMC
Bioinformatics, 17, 411-416.