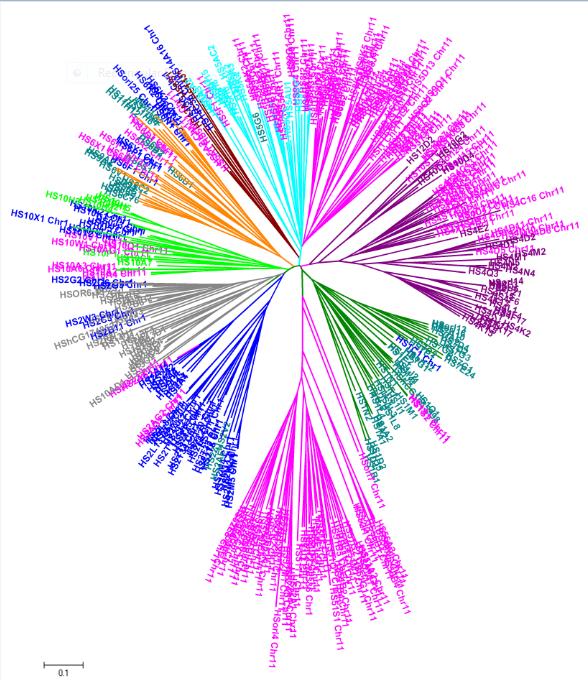
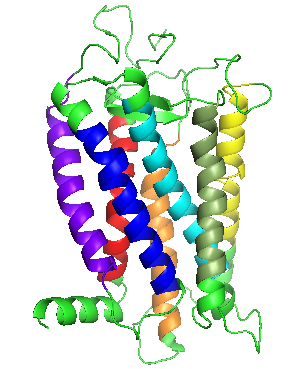
DOR is a database that provides sequence and structural information on olfactory receptors (OR) of selected organisms (Saccharomyces cerevisiae, Drosophila melanogaster, Caenorhabditis elegans, Mus musculus and Homo sapiens). Olfactory receptors are membrane proteins which help organisms to detect, encode, and process sensory stimuli. DOR provides users with olfactory receptor sequences for retrieval, predicted membrane topology, intra and inter-genomic alignments, phylogeny and identify motifs. The database contains three-dimensional structures of 100 selected ORs, modeled using bovine rhodopsin as template. The user can browse through the alignment used for comparative modeling of the olfactory receptors, structural models of the receptors, analyse the transmembrane regions marked on the model and the predicted dimer interface of every model.
Last Updated: 30-4-2015
Contributors: B.Nagarathnam, K.Harini, Kannan Sankar, Mohammed Iftekhar, D. Rajesh, S.Gigi, G. Archunan, V.Balakrishnan, M. Gromiha, W. Nemoto, K. Fukui and R.Sowdhamini
PLEASE CITE: B. Nagarathnam, Snehal D. Karpe, K. Harini, Kannan Sankar, Mohammed Iftekar, D. Rajesh, S. Giji, G. Archunan, V. Balakrishnan, M. Gromiha, W. Nemoto, K. Fukui and R. Sowdhamini. (2014) DOR- A Database of Olfactory Receptors- integrated repository for sequence and secondary structural information of olfactory receptors in selected eukaryotic genomes. Bioinformatics and Biology Insights 2014:8 147-158 .