HELP | 3PFDB+ - Database of Best Representative Profilies of Protein Families
3PFDB+ database is designed to find a best representative sequence (BRS) for each PFAM family . A profile generated from the best representative sequence, (best representative profile, BRP) is expected to identify maximum number of family members. These profiles can provide a more refined representation, owing to the large diversity observed in certain families. Sensitive homology detection methods like HMMER and FASSM were used for associate new sequences with the representative family profiles. Repertoire of representative family sequences can be used to carry out simple sequence searches which are computationally very fast and they also serve as targets for structure determination or computational modeling.
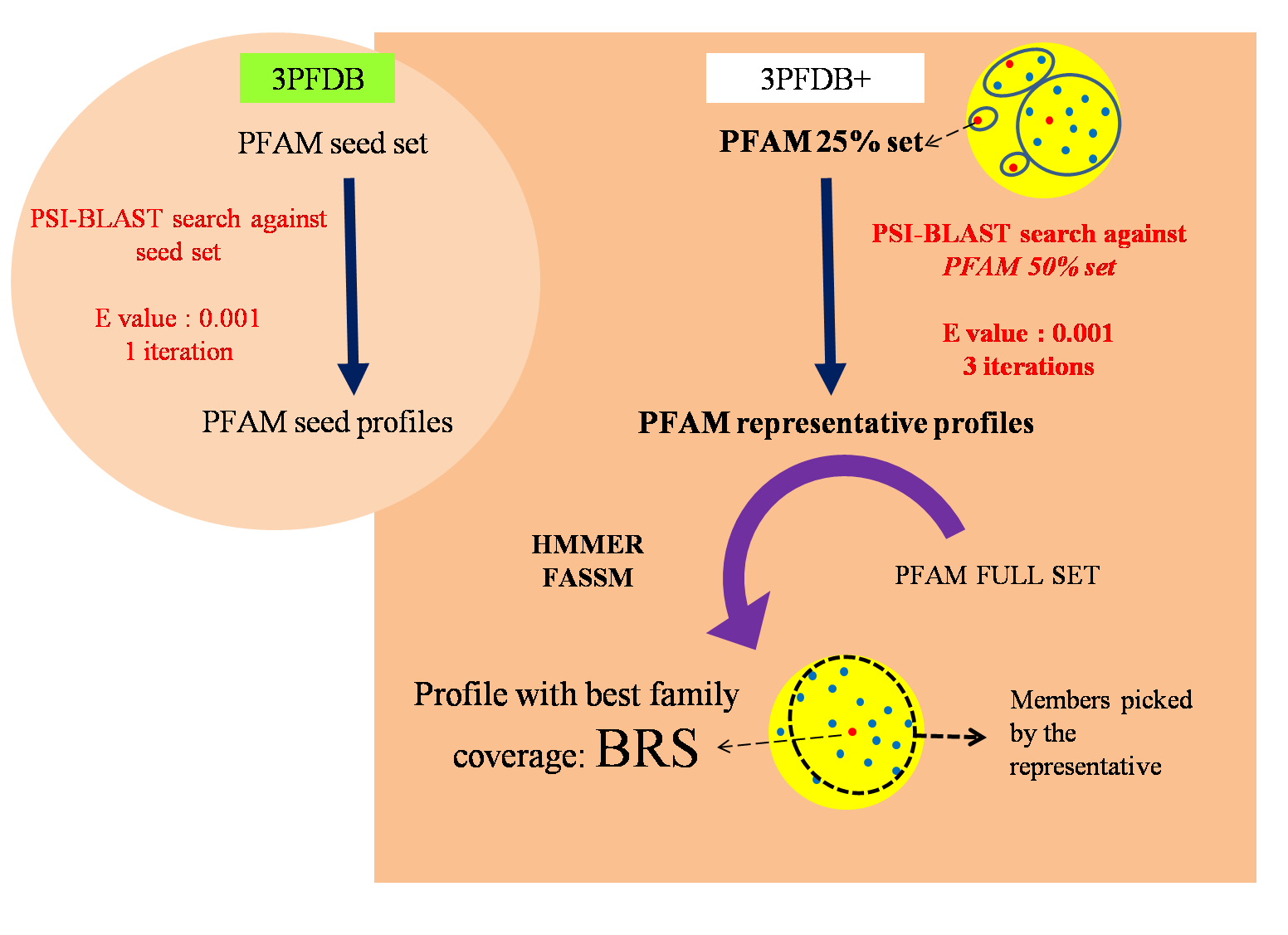
Methodology:
To extract the set of possible representatives, all the family members were clustered at a sequence identity threshold of 25%. Profiles corresponding to the representative sequences were generated by performing PSI-BLAST searches against a non-redundant PFAM family dataset gathered at 50% identity cut-off. Each of these profiles were then assessed for the family coverage using HMMER and FASSM. The efficiency in identifying other members from the same PFAM family was computed as the family coverage.
PSSM for Pfam families are selected using Coverage Analysis of FASSM Score:
A detailed explanation of FASSM method is available here - FASSM: Enhanced Function Association in whole genome analysis using Sequence and Structural Motifs In Silico Biol. 5, 425-38. Individual sequences from Pfam families are used to search against the family itself at a relaxed E-Value of 10 using PSI-BLAST. PSI-BLAST based alignments are used to generate PSSM profiles for each entries.
3PFDB+ - Database Content: In the Details section of a 3PFDB+ entry following details are available.
- PFam ID
- PFam Domain Description
- Type of sequence used for Profile Generation (Full length Sequence / Only Domain sequence)
- Number of Sequences in 50% dataset
- Pairwise Identity of Sequences in 50% dataset
- FASSM based Coverage Analysis Results
- PSIMOT-Motifs extracted using PSIMOT routine of FASSM
- Sequence based PCA plot of the Protein Famly
- Alignment of Protein Family using the sequence best representative sequence as the pilot sequence
- Download PSSM, HMM Model, Alignment and FASSM coverage analysis files
Coverage Analysis:A family sequence subset filtered at a sequence identity cut-off of less than 25% was used as the query set. Instead of using PFAM seeds for generating representative profiles, independent sequence sets gathered with an identity threshold of 50%, were used. For these representative profiles, the efficiency in identifying other members from the same PFAM family was computed as the family coverage. FASSM program incorporates different parameters, viz., size of the motif, number of the motifs allowed in the family, order of the motifs, distance between motifs, motif conservation score, etc. FASSM program employs neural network with optimized weights for each parameter to decide whether query sequence belongs to given PFAM family. The coverage for each seed sequence of PFAM family was calculated and that seed with maximum coverage were considered as BRS. 3PFDB+ generated in this study, provides representative sequences and profiles for PFAM families, with 13519 family representatives having more than 90% family coverage.
Pfam Families not included in the current version of 3PFDB+ :
PF13439, PF07690, PF03991, PF02518, PF01040, PF00353
These families were not included as they failed in generating PSSM profiles either due to short sequence length or large family sizes.
3PFDB+ - Database Features:
- FASSM based Coverage Analysis Results
- Details about the best representative member of the Family
- PSIMOT-Motifs extracted using PSIMOT routine of FASSM
- Sequence based PCA plot of the Protein Famly
- Alignment of Protein Family
- Download PSSM, HMM Model and alignment
- Details about PFam Families
Applications of 3PFDB+:
Availability of wide collection of family specific PSSMs created using best representative reference sequence will be useful for large scale protein family analysis.
- PSSMs are commonly used in the detection of remote homologues
- PSSM Profiles from 3PFDB can be used as input in NCBI-BLAST
- PSSMs corresponding to a given family can be used for in a RPS_BLAST search
- Machine Learning - PSSM can be used as an effective feature in pattern recognition
- PSI-MOT based motifs will be useful as a finger-print of protein families
3PFDB+ Team :
Prof. R. Sowdhamini (Contact : mini@ncbs.res.in)
Agnel P. Joseph, Prashant Shingate and Atul K Upadhyay
