|
DSDBASE is a database on disulphide bonds in proteins that provides information on native disulphides and those which are stereochemically possible between pairs of residues in a protein. One of the potential uses of such a disulphide database is to design site-directed mutants in order to enhance the thermal stability of the protein in question. Another important application is to employ this database for proposing 3D models of disulphide-rich polypeptides like toxins and small proteins by searching for sub-structural motifs which are compatible with the derived disulphide bond connectivity. The user can obtain information about disulphide bonds of a particular protein of interest or probe the database for multiple disulphide bonded systems of particular connectivity. The DSDBASE database has not only been updated to include 153,944 PDB entries, 216,096 native and 20,153,850 modelled disulphide bond segments, the current database provides resource to user-friendly search for multiple disulphide bond containing loops, along with annotation of their function using GO and active site residues. Further, it is now also possible to obtain three-dimensional models of disulphide-rich small proteins using an independent algorithm, RANMOD (Sowdhamini et al., 1993)that generates and examines random, but allowed backbone conformations to the polypeptide. NEW FEATURES
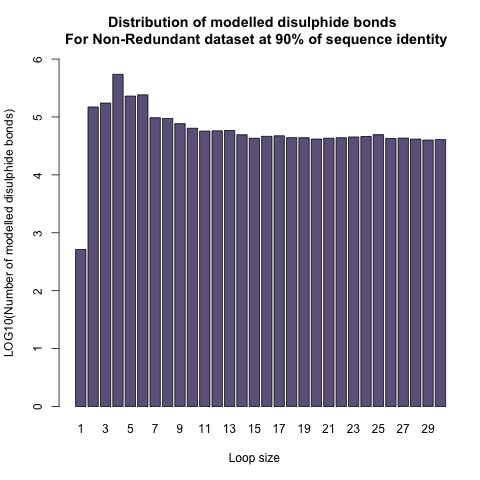
Distribution of Disulphide Bonds in DSDBASE Version 2.0
Distribution of Disulphide Bonds in DSDBASE Old Versions
|

|