There are three tabs
Title of the Run : Specify the title of the run. This is displayed in the results as a tile of the RANMOD run.
Number of Residues : number of amino acids in a given polypeptide.
Seed Number : This can be any number in integer and determines the choice of random numbers and hence the conformation generated. The result of a run can be reproduced using this number.
Number of Runs : Number of conformations (NRUN) to be searched.
Disulphide Connections : This is for information on disulphides. Within this field, the residue number of Cys (i) and Cys(j) (where Cys(i)-Cys(j) is a SS bond) is provided through specifying the start residue(i) and end residue(j) of the connection in the number input box. Add button adds the connections one after the other. Remove button helps in removing connections entered incorrectly.
Helix List : This field is for additional information if present on helices. For a helix, the residue number where the helix begins and the residue number where the helix ends are given is provided through specifying the start residue and end residue of the helix in the number input box. Add button adds the connections one after the other. Remove button helps in removing connections entered incorrectly.
Strand List : This is similar to field on helix list but for extended strands (IE).
Glycyl List : This field is for glycines. The residue number of the glycine is specified through number input box and add button.
Prolyl List : This field is for prolines. The residue number of the glycine is specified through number input box and add button.
CIS Peptide List : The residue number of X (for a situation where a cis-peptide is present at the peptide connecting X and Y; X-Y) and the percentage of cis-conformation (in 0-100 range) are provided through the number input box. Add button adds to the list one after the other. Remove button helps in removing connections entered incorrectly.
Sequence Text for PDB model : The amino acid sequence(FASTA format without the header) example : CSCSSLMDKECVYFCHL. This is used for generating PDB (getPDB option in the RANMOD output screen) from the models generated by RANMOD.
OR
Sequence File for PDB model : The above information provided in a file which can be uploaded.
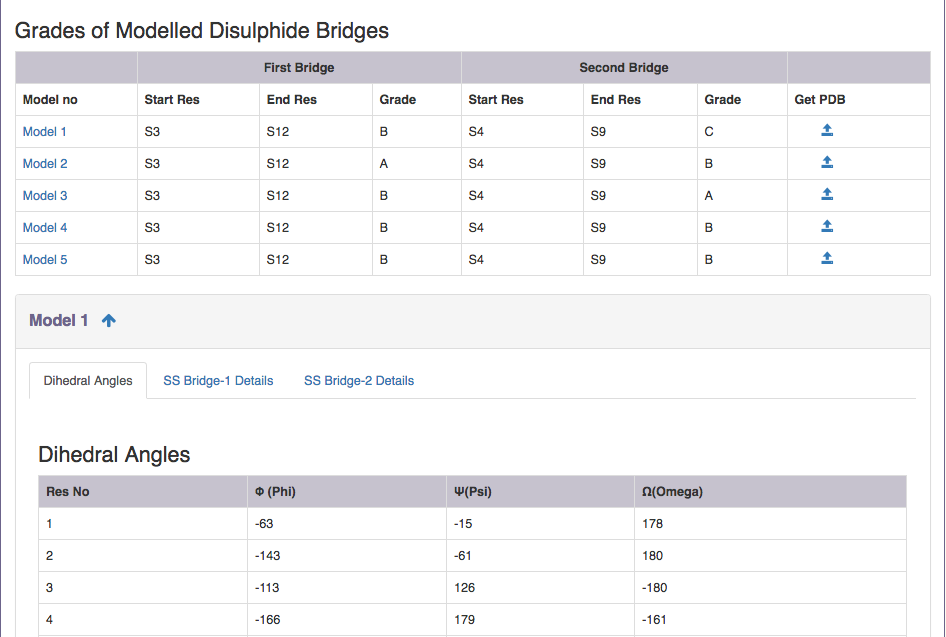
| Output features | Description |
|---|---|
| Model no | Takes you to the respective model information. |
| Start Res | Gives information on the start residue of the disulphide bridge. |
| End Res | Gives information on the start residue of the disulphide bridge. |
| Grade | Grade distribution of modelled disulphides. |
| Get PDB | Gives PDB file of the modelled peptide ( Example: for Model 1, gives PDB file of the modelled peptide ). User can view the segment in Jsmol and PDB file is available for download. Backbone of all residues are fixed by RANMOD.CYS residues part of the disulphide loop connectivity are fixed by RANMOD and side chain of all other residues are fixed by SCWRL package of Dunbrack Lab. |
(for more explanation on the PDB file, refer to DSDBASE central help page from here. )
Following this, for each model, results have 3 tabs
Dihedral Angles :A table gives the information about the various backbone torsional angles such as phi, psi and omega at every amino acid residues in degrees.
SS Bridge-1 Details :a table on sulphur coordinates of SS bridge 1 and SS parameters are displayed.
SS Bridge-2Details: a table on sulphur coordinates of SS bridge 2 and SS parameters are displayed.
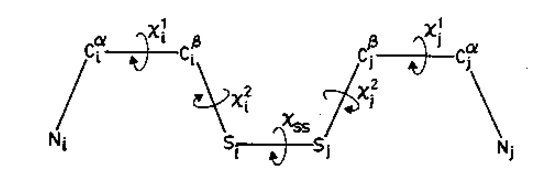
Figure. Various dihedral angles in a cysteine disulphide.
A general procedure for arriving at 3-D models of disulphide-rich polypeptide systems based on the covalent cross-link constraints. The procedure, RANMOD, assigns a large number of random, permitted backbone conformations to the polypeptide and identifies stereochemically acceptable structures as plausible models based on strainless disulphide bridge modelling. Disulphide bond modelling is performed using the procedure MODIP (MOdelling of DIsulphide bonds in Proteins).
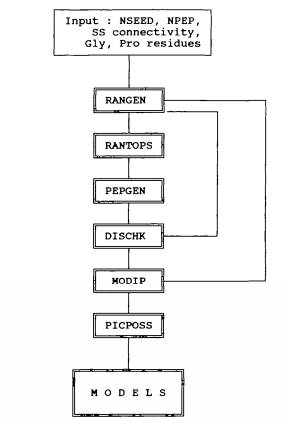
Flow chart of the RANMOD procedure used for modelling polypeptides rich in disulphide bonds. The required inputs include an integer for defining the seed number for random-number generation, NSEED, the number of residues in the peptide of interest, NPEP and information about the disulphide bond connectivity and the location of Gly and Pro residues (if any). The subroutine RANGEN generates random numbers while RANTOPS converts the random numbers to the (Phi, Psi) scale. Assignment of the backbone torsion angles at each residue of the peptide is done within the permitted (Phi, Psi) space depending on the nature of the residue, whether, it is Gly, Pro or Non-Gly-Non-Pro. PEPGEN generates the conformation of the peptide corresponding to the random backbone conformation assignment. The two distance constraints corresponding to C(alpha)-C(alpha) and C(beta)-C(beta) at every bridge position are applied by the subroutine DISCHK. Those conformations which conform to the disulphide distance constraints are examined for stereochemical suitability by the subroutine MODIP. PICPOS selects best positions for the disulphide bridges modelled by MODIP.
R.Sowdhamini, N.Srinivasan, B.Shoichet, D.V.Santi, C.Ramakrishnan and P.Balaram (1989). Stereochemical modelling of disulfide bridges: Criteria for introduction into proteins by site-directed mutagenesis. Prot. Engng., 3, 95-103.
R.Sowdhamini, C.Ramakrishnan and P.Balaram (1993). Modelling multiple disulphide loop containing polypeptides by random conformation generation. The test cases of alpha-conotoxin GI and endothelin I. Protein Eng. ,6(8):873-82.
G. G. Krivov, M. V. Shapovalov, and R. L. Dunbrack, Jr. Improved prediction of protein side-chain conformations with SCWRL4. Proteins (2009).