There are three tabs
Protein name: Enter any name (this is optional)
Database name: Currently four different types of datasets are available for DSDBASE search.
| Select Database | Description |
|---|---|
| nr-db-30% | a database of native and modelled disulphide bonds derived from non-redundant set of proteins (30% sequence identity) derived from PDB(Jan 2021). |
| nr-native-30% | a database of only native disulphide bonds derived from non-redundant set of proteins (30% sequence identity) derived from PDB (Jan 2021). |
| nr-db-90% | a database of native and modelled disulphide bonds derived from non-redundant set of proteins (90% sequence identity) derived from PDB(Jan 2021). |
| fulldb (PDB-Jan 2021) | a database of native and modelled disulphide bonds derived from full PDB release (May-2016), |
Disulphide bond connectivity: This is for information on disulphides. Within this field, the residue number of Cys (i) and Cys(j) (where Cys(i)-Cys(j) is a SS bond) is provided through specifying the start residue(i) and end residue(j) of the connection in the number input box. Add button adds the connections one after the other. Remove button helps in removing connections entered incorrectly.
Advance Search Options
a. Explanation of loopsize relaxation
Now we can represent our connectivity pattern as follows
Loop 1:

In this case loop size = ( 15-1 ) + 1 = 15
If loop size is relaxed by 1 residue then search can be performed for loop size 15, 14 (15-1), 16 (15+1)
Loop 2:
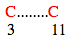
In this case loop size = ( 11-3 ) + 1 = 9
If loop size is relaxed by 1 residue then search can be performed for loop size 9, 8 (9-1), 10(9+1)
Loop proximity of given query can be represent like this

Spatial distance between starting residue of bond1 and bond2 = 3 - 1 = +1
Spatial distance between end residue of bond1 and bond2 = 11 -15 = -3
By default (even without invoking this option), the search engine looks for any sub-structural motif of a protein having loop sizes similar to query and also their spatial distances.
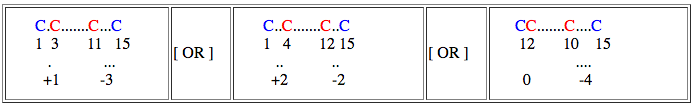
If spacial distance is relaxed by 1 residue then,
spatial distance for starting residue : +1, 0 (1-1), 2 (1+1)
spatial distance for end residue : -3, -4 (-3-1), -2 (-3+1)
and search can be performed as follows,
5. Sequence Information: This is optional. Enter the sequence of the protein/peptide. Please note that only the portion of protein/prptide whose connectivity is being searched is to be provided. In this example "CSCSSLMDKECVYFC" is to be pasted into the text area(the full length sequence is "CSCSSLMDKECVYFCHL". Donot include charactres other than alphabets. No particular format is needed. Simply paste the raw sequence.
The output file contains following informations
Screen shot of search result for SS connectivity 1,15 3,11
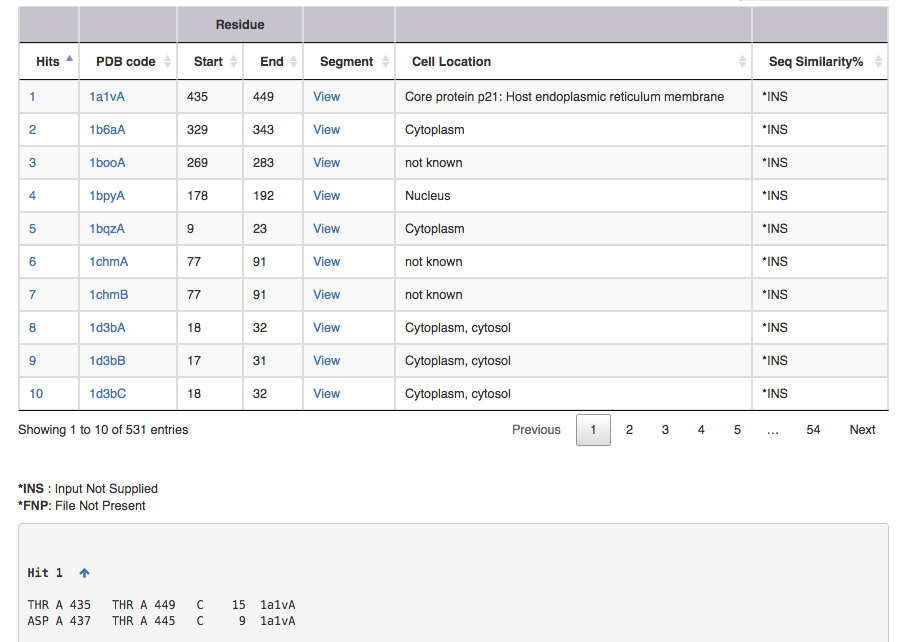
| Output features | Description |
|---|---|
| Hit | takes you to the substructure information (see below for more on this) |
| PDB code | leads to the corresponding entry in DSDBASE (for more information, click [here]. |
| Segment | gives PDB file for given region alone ( Example: for Hit 1 segment gives PDB file for residue no 15 to 29 ). User can view the segment in Jsmol and PDB file is available for download. |
| CelLoc | CelLoc is the subcellular location of the protein. The results are extracted from PDB file. "Not Known" - SubLoc couldn't predict the cellular location of the protein. |
| SeqSim | SeqSim is the the percentage of sequence similarity of the click with the query sequence. Note that the cysteine positions are not fixed. INS = Input Not Supplied. FNP = File Not Present: Most probably this may be an obsolete/theoretical entry in the PDB. |
A hit is ...
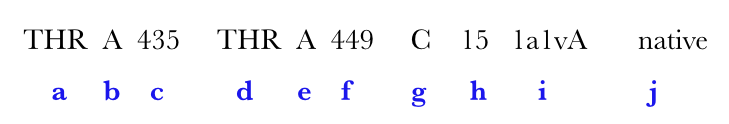
| Output features | Description |
|---|---|
| a | amino acid in ith,position |
| b | chain identifier |
| c | amino acid number of ith position |
| d | amino acid in jth position |
| e | chain identifier |
| f | amino acid number of jth position |
| g | grade |
| h | loopsize |
| i | PDB code |
| j (optional) | Native can be represented in either blue or red. (for explanation see below) |
Native represents a substructural connectivity where both the residues are CYS(Cysteine). In most instances these are as annotated in the PDB file and here represented in Blue color. If MODIP only predicts these connectivities, they are represented in Red color.
Redox-active represents redox-active disulphide bonds which are functionally important and expected to behave differently from typical structural disulphide bonds. These are reductive, reaction probably involves nucleophilic attack by the Cys thiolate on the substrate to form a mixed disulphide intermediate.
The important application is to employ DSDBASE for proposing 3D models of disulphide-rich polypetides like toxins and small proteins.such small proteins are not rich in secondary structure and are stabilised by the covalent crosslinks. This database provides information about all possible positions to introduce disulphide bonds in proteins along with there stereochemical grades. Option to get mutant file of interest and graphical view of mutant pdb is also available.
R.Sowdhamini, N.Srinivasan, B.Shoichet, D.V.Santi, C.Ramakrishnan and P.Balaram (1989). Stereochemical modelling of disulfide bridges: Criteria for introduction into proteins by site-directed mutagenesis. Prot. Engng., 3, 95-103.
Thangudu, R. R., Vinayagam, A., Pugalenthi, G.,Manonmani, A., Offmann, B. & Sowdhamini, R. (2005) Native and modelled disulphide bonds in proteins: Knowledge-based approaches towards structure prediction of disulphide-rich polypeptides. Proteins: Structure Function and Bioinformatics, 58, 866-79.