DSDBASE Central Help |
This disulphide database is useful to design site-directed mutants in order to enhance the thermal stability of the proteins in question. In this database disulphide bonds were designed in the stereochemicaly optimal region of the protein, so one can introduce a disulphide bond without straining the protein using this database.
This database provides information about all possible positions to introduce disulphide bonds in proteins along with there stereochemical grades. Option to get mutant file of interest and graphical view of mutant PDB is also available. Present release considers PDB Jan 2021 release.
The browse option in top level menu contains information about the following:
All entries : contains list of PDB entries from the present release. Each PDB hit takes them to the relevant page containing information on disulphide connectivity.
Disulphide Peptides : User can download peptide fragments of distinct loop sizes from various substructures as a zipped file.
The enzymes option in top level menu contains information about the six classes of enzymes as follows:
User can go to a PDB hit belonging to specific class of enzymes. Further, a click on Hit from PDB, will take them to a page containing information on disulphide connectivity.
Using the search panel on top right, user can search database using the following options:
Search by PDB code - To search by PDB code, enter a valid PDB code (4 letter code starts with number). The result page contains PDB code (with link) and Protein name. User can reach the choosen entry by using link provided in PDB code.
Search by EC(Enzyme Commission) Number - To search by EC Number, enter a valid EC number (separated by period).The result page contains PDB code (with link) ,EC number and Protein name. User can reach the choosen entry by using link provided in PDB code.
You may type 1 digit into the first field (the enzyme class), 1 or 2 digits into the second and third fields (subclass and sub-subclass), and up to 3 digits into the fourth field. For example, you may type 1.2.1.26 to get the corresponding ENZYME entry.You may also enter a partial EC number in order to get a list of all ENZYME entries whose EC numbers begin with the given pattern. For example, you may just type 1.2
Search by Keyword - To search by keyword, enter keyword. The result page contains PDB code (with link) and Protein name. User can reach the choosen entry by using link provided in PDB code.
Search is available from all the pages. Other than this, for new or missing entries option to run MODIP online also available.
The important application is to employ database for proposing three- dimensional models of disulphide-rich polypetides like toxins and small proteins.such small proteins are not rich in secondary structure and are stabilised by the covalent crosslinks.
Using tools option in top level menu, user can run one of the three modelling procedures.
MODIP : Disulphide details for Missing entries or New PDB files can obtained by running MODIP program online.
RANMOD: Assigns plausible models to peptides that are rich in disulphides bonds based on the stereochemistry of the covalent crosslinks.
Modelling peptides : One can probe the database for multiple disulphide bond systems of particular connectivity and obtain the structure of the Protein.
There are three tabs
None
Results have 2 tabs
Summary : This gives information on the number of native and modelled disulphides and distribution of grades.
Disulphide-Details : Native and Modelled disulphide details in the form of table.
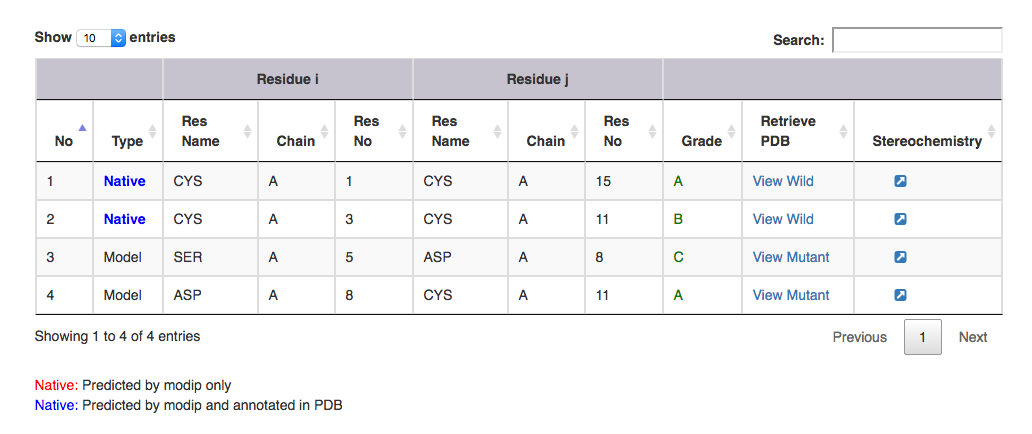
| Output features | Description |
|---|---|
| Type | gives information on whether it is a modelled or a native disulphide bridge. Annotated and unannotated SS bonds: where both the residues are cysteines. If marked in the PDB file, blue colour. If predicted by MODIP then native is represented in red colour. |
| Residue i | gives information on the start residue name, chain ID and residue number of the SS bond. |
| Residue j | gives information on the end residue name, chain ID and residue number of the SS bond. |
| Grade | grade distribution of modelled and native disulphides. |
| Retrive PDB | gives PDB file of the substructural segment with residue i and j mutated to cysteines ( Example: for Modelled SS bond, mutant PDB file is available with residue no 5 to 8 mutated to Cysteines ). User can view the segment in Jsmol and PDB file is available for download. |
| Stereochemistry | hyperlink to view the stereochemical parameters of SS bond. |
There are three tabs
Title of the Run : Specify the title of the run. This is displayed in the results as a tile of the RANMOD run.
Number of Residues : number of amino acids in a given polypeptide.
Seed Number : This can be any number in integer and determines the choice of random numbers and hence the conformation generated. The result of a run can be reproduced using this number.
Number of Runs : Number of conformations (NRUN) to be searched.
Disulphide Connections : This is for information on disulphides. Within this field, the residue number of Cys (i) and Cys(j) (where Cys(i)-Cys(j) is a SS bond) is given in comma separated. This is followed by as many spaces as there are disulphides. For example, 3,10 9,15 implies first disulphide bond is between the residue number 3 and 10, and the second disulphide bond is between the residue number 9 and 15, respectively.
Helix List : This field is for additional information if present on helices. This is followed by as many spaces as there are helices predicted. For a helix, the residue number where the helix begins and the residue number where the helix ends are given in comma separated format.
Strand List : This is similar to field on helix list but for extended strands (IE).
Glycyl List : This field is for glycines. The residue number of the glycine is given in space seperated format.
Prolyl List : This field is for prolines. The residue number of the proline is given in space seperated format.
CIS Peptide List : The residue number of X (for a situation where a cis-peptide is present at the peptide connecting X and Y; X-Y) and the percentage of cis-conformation (in 0-100 range) are given in comma separated format. This is followed by as many spaces as there are cis peptides.
Sequence Text for PDB model : The amino acid sequence(FASTA format without the header) example : CSCSSLMDKECVYFCHL. This is used for generating PDB (getPDB option in the RANMOD output screen) from the models generated by RANMOD.
OR
Sequence File for PDB model : The above information provided in a file which can be uploaded.
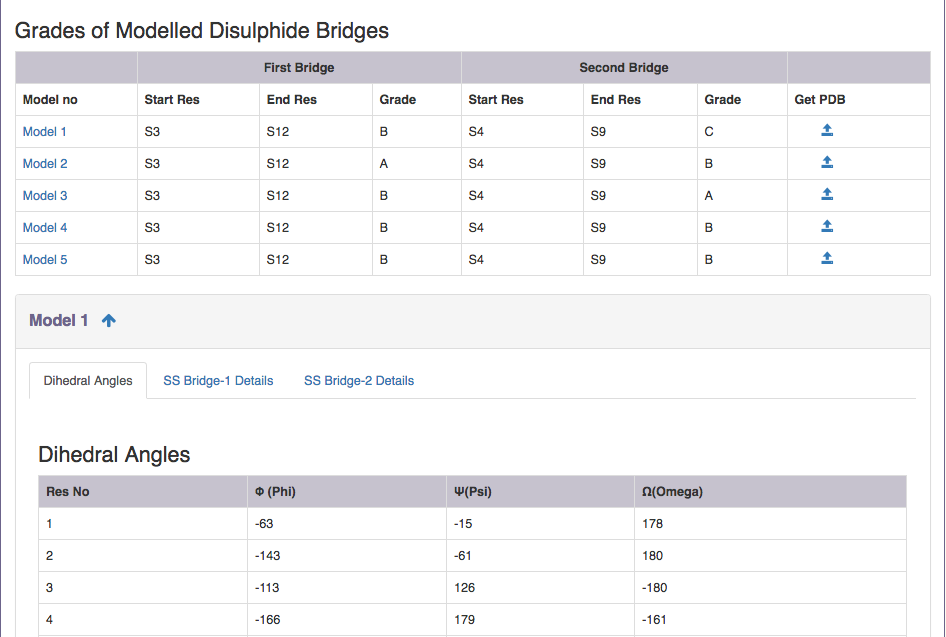
| Output features | Description |
|---|---|
| Model no | Takes you to the respective model information. |
| Start Res | Gives information on the start residue of the disulphide bridge. |
| End Res | Gives information on the start residue of the disulphide bridge. |
| Grade | Grade distribution of modelled disulphides. |
| Get PDB | Gives PDB file of the modelled peptide ( Example: for Model 1, gives PDB file of the modelled peptide ). User can view the segment in Jsmol and PDB file is available for download. Backbone of all residues are fixed by RANMOD.CYS residues part of the disulphide loop connectivity are fixed by RANMOD and side chain of all other residues are fixed by SCWRL package of Dunbrack Lab. |
The output contains the co-ordinates in the PDB format. In case of Cys, SG positions of the Cys residues are provided by RANMOD. The backbone of the entire residue chain as provided in the input form is fixed by RANMOD.The side chain of all other residues are fixed by SCWRL package of Dunbrack Lab.
By the very nature of the procedure, multiple conformationsare proposed as 3D models of the polypeptide. It is upto the user to identify the "best" ones. The present version does not rank the proposed models.
Following this, for each model, results have 3 tabs
Dihedral Angles :A table gives the information about the various backbone torsional angles such as phi, psi and omega at every amino acid residues in degrees.
SS Bridge-1 Details :a table on sulphur coordinates of SS bridge 1 and SS parameters are displayed.
SS Bridge-2Details: a table on sulphur coordinates of SS bridge 2 and SS parameters are displayed.
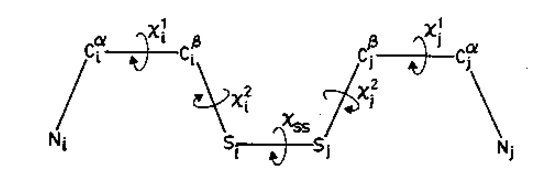
Figure. Various dihedral angles in a cysteine disulphide.
There are three tabs
Protein name: Enter any name (this is optional)
Database name: Currently four different types of datasets are available for DSDBASE search.
| Select Database | Description |
|---|---|
| nr-db-30% | a database of native and modelled disulphide bonds derived from non-redundant set of proteins (30% sequence identity) derived from PDB(Jan 2021). |
| nr-native-30% | a database of only native disulphide bonds derived from non-redundant set of proteins (30% sequence identity) derived from PDB (Jan 2021). |
| nr-db-90% | a database of native and modelled disulphide bonds derived from non-redundant set of proteins (90% sequence identity) derived from PDB(Jan 2021). |
| fulldb (PDB-Jan 2021) | a database of native and modelled disulphide bonds derived from full PDB release (Jan 2021), |
Disulphide bond connectivity: This is for information on disulphides. Within this field, the residue number of Cys (i) and Cys(j) (where Cys(i)-Cys(j) is a SS bond) is given in comma separated. This is followed by as many spaces as there are disulphides. For example, 3,10 9,15 implies first disulphide bond is between the residue number 3 and 10, and the second disulphide bond is between the residue number 9 and 15, respectively.
Advance Search Options
a. Explanation of loopsize relaxation
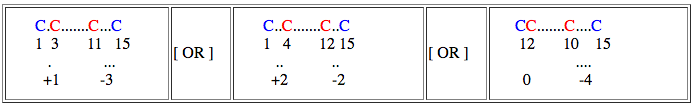
Now we can represent our connectivity pattern as follows
Loop 1:

In this case loop size = ( 15-1 ) + 1 = 15
If loop size is relaxed by 1 residue then search can be performed for loop size 15, 14 (15-1), 16 (15+1)
Loop 2:
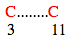
In this case loop size = ( 11-3 ) + 1 = 9
If loop size is relaxed by 1 residue then search can be performed for loop size 9, 8 (9-1), 10(9+1)
Loop proximity of given query can be represent like this

Spatial distance between starting residue of bond1 and bond2 = 3 - 1 = +1
Spatial distance between end residue of bond1 and bond2 = 11 -15 = -3
By default (even without invoking this option), the search engine looks for any sub-structural motif of a protein having loop sizes similar to query and also their spatial distances.
If spacial distance is relaxed by 1 residue then,
spatial distance for starting residue : +1, 0 (1-1), 2 (1+1)
spatial distance for end residue : -3, -4 (-3-1), -2 (-3+1)
and search can be performed as follows,
5. Sequence Information: This is optional. Enter the sequence of the protein/peptide. Please note that only the portion of protein/prptide whose connectivity is being searched is to be provided. In this example "CSCSSLMDKECVYFC" is to be pasted into the text area(the full length sequence is "CSCSSLMDKECVYFCHL". Donot include charactres other than alphabets. No particular format is needed. Simply paste the raw sequence.
The output file contains following informations
Screen shot of search result for SS connectivity 1,15 3,11
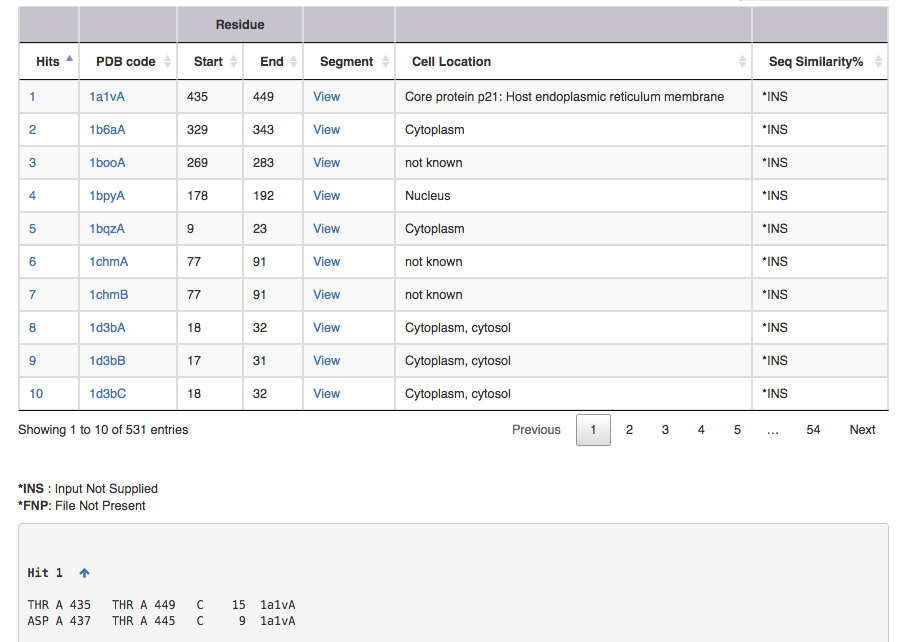
| Output features | Description |
|---|---|
| Hit | takes you to the substructure information (see below for more on this) |
| PDB code | leads to the corresponding entry in DSDBASE (for more information, click [here]. |
| Segment | gives PDB file for given region alone ( Example: for Hit 1 segment gives PDB file for residue no 15 to 29 ). User can view the segment in Jsmol and PDB file is available for download. |
| CelLoc | CelLoc is the subcellular location of the protein. The results are extracted from PDB file. "Not Known" - SubLoc couldn't predict the cellular location of the protein. |
| SeqSim | SeqSim is the the percentage of sequence similarity of the click with the query sequence. Note that the cysteine positions are not fixed. INS = Input Not Supplied. FNP = File Not Present: Most probably this may be an obsolete/theoretical entry in the PDB. |
A hit is ...
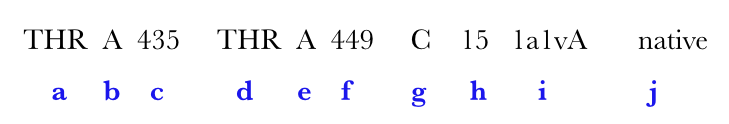
| Output features | Description |
|---|---|
| a | amino acid in ith,position |
| b | chain identifier |
| c | amino acid number of ith position |
| d | amino acid in jth position |
| e | chain identifier |
| f | amino acid number of jth position |
| g | grade |
| h | loopsize |
| i | PDB code |
| j (optional) | Native can be represented in either blue or red. (for explanation see below) |
Native represents a substructural connectivity where both the residues are CYS(Cysteine). In most instances these are as annotated in the PDB file and here represented in Blue color. If MODIP only predicts these connectivities, they are represented in Red color.
Redox-active represents redox-active disulphide bonds which are functionally important and expected to behave differently from typical structural disulphide bonds. These are reductive, reaction probably involves nucleophilic attack by the Cys thiolate on the substrate to form a mixed disulphide intermediate.
R.Sowdhamini, N.Srinivasan, B.Shoichet, D.V.Santi, C.Ramakrishnan and P.Balaram (1989). Stereochemical modelling of disulfide bridges: Criteria for introduction into proteins by site-directed mutagenesis. Prot. Engng., 3, 95-103.
N.Srinivasan, R.Sowdhamini, C.Ramakrishnan and P.Balaram (1990). Conformations of disulfide bridges in proteins and peptides. Int. J. Pept. Prot. Res.,36, 147-155.
R.Sowdhamini, C.Ramakrishnan and P.Balaram (1993). Modelling multiple disulphide loop containing polypeptides by random conformation generation. The test cases of alpha-conotoxin GI and endothelin I. Protein Eng. ,6(8):873-82.
L.E.Donate, E.Gherardi, N.Srinivasan, R.Sowdhamini, S.Aparicio and T.L.Blundell (1994). Molecular evolution and domain structure of plasminogen-related growth factors (HGF/SF and HGFl/MSP). Prot. Sci., 3, 2378-2394.
H.Zhou, M.J.Mazzulla, J.D.Kaufman, S.J.Stahl, P.T.Wingfield, J.S.Rubin, D.P.Bottaro and R.A.Byrd (1998). The solution structure of the N-terminal domain of hepatocyte growth factor reveals a potential heparin-binding site. Structure., 6(1), 109-16.
D.Y.Chirgadze, J.Hepple, R.A.Byrd, R.Sowdhamini and T.L.Blundell (1998). Insights into the structure of hepatocyte growth factor scatter factor (HGF/SF) and implications for receptor activation. FEBS Letts., 430, 126-129.
R.R.Thangudu, A.Vinayagam, G.Pugalenthi, A.Manonmani, B.Offmann and R.Sowdhamini (2005). Native and modelled disulphide bonds in proteins: Knowledge-based approaches towards structure prediction of disulphide-rich polypeptides. Proteins: Struct., Funct., Bioinf., 58, 866-79.
G. G. Krivov, M. V. Shapovalov, and R. L. Dunbrack, Jr. Improved prediction of protein side-chain conformations with SCWRL4. Proteins (2009).