3DSwap+ - Semi-automated Knowledgebase of 3D domain swapping:
3DSwap+ is a comprehensive collection of protein structures that are reported to be involved in 3-Dimensional Domain Swapping (3DSwap). 3DSwap+ database contains data at various level: sequence (interface and hinge region), structure (for graphics visualization, interface regions, objective measures for extent of swapping, features such as hydrogen bonding, solvent accessibility and backbone conformation) and function (annotations from GO and Pfam). 3D domain swapping is a mechanism for two or more protein molecules to form a dimer or higher oligomer by exchanging an identical structural element known as domain. It has been first reported in Diptheria Toxin. 3DSwap+ provides detailed information about swapping, protein-protein interface, extent of swapping etc. Information related to domain swapping were extracted by HEIDI, maually and also from orginal research publications after extensive literature curation.
3D Domain Swapping : Structural features:
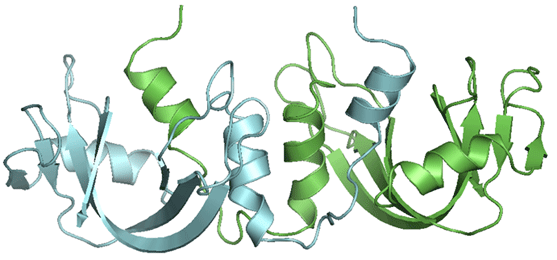
3D Domain Swapping : This structural phenomenon is observed in a large number of structures, where two or more chains form an oligomeric structure by exchanging a structural segment between the chains.
Hinge Region : The residues(or short sequence) which connects the secondary minor and major region is called as hinge region. The hinge region can be a loop, sheet, helix or few residues.
Swapped region : A globular domain (or sometimes one or few elements of secondary structure) that is intertwined with an identical protein chain, is referred as the swapped segment. The swapped domain retains an environment essentially identical to that of the same domain in a protein monomer.
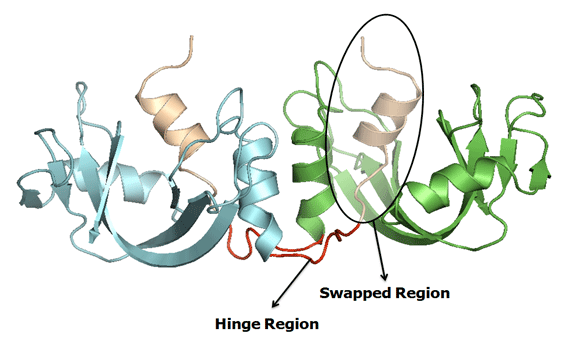
Secondary Minor Region : The entire domain or a part of one chain which is swapped with the other domain of the neighbouring chain is referred as "secondary minor region" or Guest. Secondary minor region is an alternate definition used to define hinge region to explain the secondary regions with more clarity.
Secondary Major Region * : The region which interacts with the secondary minor region is termed as Secondary major region or Host.
Quasi-Domain Swapping : Quasidomain swapping refers to the structures where some proteins form domain swapped oligomers without a known closed monomer. If these proteins have homologues known to be closed monomers, these oligomers are considered to be .quasidomain swapped. Examples of quasidomain swapping include crystallin, pheromone/odorant binding/transport proteins, RYMV (viral capsid protein) , human cystatin C (protease inhibitor) etc.
Bonafide Domain Swapping : Bonafide domain swapping refers to structures where both the monomer and the dimer of a molecule exist in stable forms, where the dimer adopts a domain-swapped conformation and the monomer adopts a closed conformation. Examples of Bonafide domain swapping includes diphtheria toxin, RNases, CD2 (T-cell adhesion), Spo0A (development regulation) etc.
Extent of swapping * : Extent of swapping is a new method developed to assess of the extent of swapping with respect to the protein chains involved in swapping. For example, for a protein with 2 chains X, Y, the extent of swapping is calculated using the formula given below:
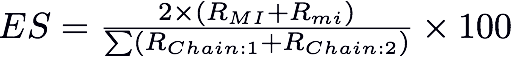
Where RMI= Number of residues in secondary major interface, Rmi = Number of residues in secondary minor interface. RChain:1 = Number of residues in Chain 1 and RChain:2 = Number of residues in Chain 2. Based on the percentage of extent of swapping observed in different structures, we have classified the structures into three major classes as 'Extensive swapping', 'Moderate swapping' and 'Minimum swapping'. Extensive swapping refers to the oligomeric structures where the 'Extent of Swapping' is observed within the range of 35%-100%. Moderate swapping refers to the oligomeric structures where the 'Extent of Swapping' is observed within the range of 15%-35%. Minimum swapping refers to the oligomeric structures where the 'Extent of Swapping' is observed within the range of 1-15%.
3DSwap+ - Protein Structural Curation Method:
Protein structures presented in 3DSwap+ was curated using a new curation approach i.e., HEIDI which idendifies hinge regions based on numer of inter and intramolecular interactions. Hinge region in domain-swapped proteins acquires extended conformation and hence, hinge region contains fewer intramolecular interactions, moderate to high intermolecular interactions, is positioned far from the core of the parent subunit, is solvent-accessible and retains extended backbone conformation. We combined literature mining approach and manual structural analysis and visualization in an iterative fashion to identify hinge and swapped regions. PDB ID, Chain ID, Oligomeric state, Hinge Regions, Swapped Region, Extent of Swapping, Sequence Data and Reference fields were curated and stored in the database.
