RANMOD - Random Conformation to Polypeptide Backbone
RANMOD - Random Conformation to Polypeptide Backbone.
There are three tabs
- User-execution : User should provide inputs for running RANMOD procedure.
- Sample-execution : Provides sample input for modelling using RANMOD procedure.
- Help : To see usage and explanation of RANMOD procedure.
Input Parameters
- Title of the Run : Specify the title of the run. This is displayed in the results as a tile of the RANMOD run.
- Number of Residues : number of amino acids in a given polypeptide.
- Seed Number : This can be any number in integer and determines the choice of random numbers and hence the conformation generated. The result of a run can be reproduced using this number.
- Number of Runs : Number of conformations (NRUN) to be searched.
- Disulphide Connections : This is for information on disulphides. Within this field, the residue number of Cys (i) and Cys(j) (where Cys(i)-Cys(j) is a SS bond) is given in comma separated. This is followed by as many spaces as there are disulphides. For example, 3,10 9,15 implies first disulphide bond is between the residue number 3 and 10, and the second disulphide bond is between the residue number 9 and 15, respectively.
- Helix List : This field is for additional information if present on helices. This is followed by as many spaces as there are helices predicted. For a helix, the residue number where the helix begins and the residue number where the helix ends are given in comma separated format.
- Strand List : This is similar to field on helix list but for extended strands (IE).
- Glycyl List : This field is for glycines. The residue number of the glycine is given in space seperated format.
- Prolyl List : This field is for prolines. The residue number of the proline is given in space seperated format.
- CIS Peptide List : The residue number of X (for a situation where a cis-peptide is present at the peptide connecting X and Y; X-Y) and the percentage of cis-conformation (in 0-100 range) are given in comma separated format. This is followed by as many spaces as there are cis peptides.
Displayed Results
- The output is displayed on to the screen in the form of a table with information such as total number of models generated, grade distribution of modelled disulphides and residue number.
- Following this, a table gives the information about the various backbone torsional angles such as phi, psi and omega at every amino acid residues in degrees.
- Further, a table on sulphur coordinates of SS bridge and SS parameters are displayed.
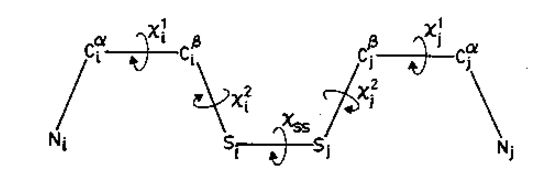
Figure. Various dihedral angles in a cysteine disulphide.
Summary of RANMOD procedure
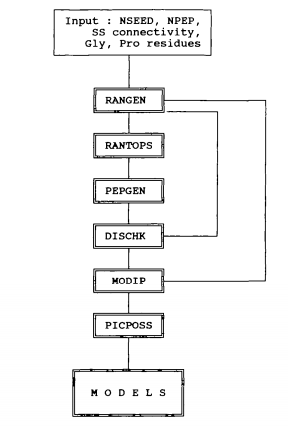
Flow chart of the RANMOD procedure used for modelling polypeptides rich in disulphide bonds. The required inputs include an integer for defining the seed number for random-number generation, NSEED, the number of residues in the peptide of interest, NPEP and information about the disulphide bond connectivity and the location of Gly and Pro residues (if any). The subroutine RANGEN generates random numbers while RANTOPS converts the random numbers to the (Phi, Psi) scale. Assignment of the backbone torsion angles at each residue of the peptide is done within the permitted (Phi, Psi) space depending on the nature of the residue, whether, it is Gly, Pro or Non-Gly-Non-Pro. PEPGEN generates the conformation of the peptide corresponding to the random backbone conformation assignment. The two distance constraints corresponding to C(alpha)-C(alpha) and C(beta)-C(beta) at every bridge position are applied by the subroutine DISCHK. Those conformations which conform to the disulphide distance constraints are examined for stereochemical suitability by the subroutine MODIP. PICPOS selects best positions for the disulphide bridges modelled by MODIP.
Downloadable Results
- Result in text format
Details
A general procedure for arriving at 3-D models of disulphide-rich polypeptide systems based on the covalent cross-link constraints. The procedure, RANMOD, assigns a large number of random, permitted backbone conformations to the polypeptide and identifies stereochemically acceptable structures as plausible models based on strainless disulphide bridge modelling. Disulphide bond modelling is performed using the procedure MODIP (MOdelling of DIsulphide bonds in Proteins).
RANMOD - Reference
Sowdhamini R, Ramakrishnan C, Balaram P. Modelling multiple disulphide loop containing polypeptides by random conformation generation. The test cases of alpha-conotoxin GI and endothelin I. Protein Eng. 1993 Nov;6(8):873-82.
Sowdhamini R, Srinivasan N, Shoichet B, Santi DV, Ramakrishnan C, Balaram P. Stereochemical modeling of disulfide bridges. Criteria for introduction into proteins by site-directed mutagenesis. Protein Eng. 1989 Nov;3(2):95-103.
Contact
Prof. R. Sowdhamini(mini@ncbs.res.in)
Sowmya Indrakumar
Murugavel P