Modip - Modelling of Disulphide bonds in proteins
Modip - Modelling of Disulphide bonds in proteins
There are three tabs
- User-execution : User should provide inputs for running MODIP procedure.
- Sample-execution : Provides sample input for modelling using MODIP procedure.
- Help : To see usage and explanation of MODIP procedure.
Input Files
PDB File : Upload File in Protein Data Bank (PDB) format- PDB file can be single chain or multi chain.
- 'C-alpha only' proteins are not useful for running MODIP.
Input Parameters
None
Displayed Results
Results have 2 tabs
Summary : This gives information on the number of native and modelled disulphides and distribution of grades.
Disulphide Details : Native and Modelled disulphide details in the form of table.
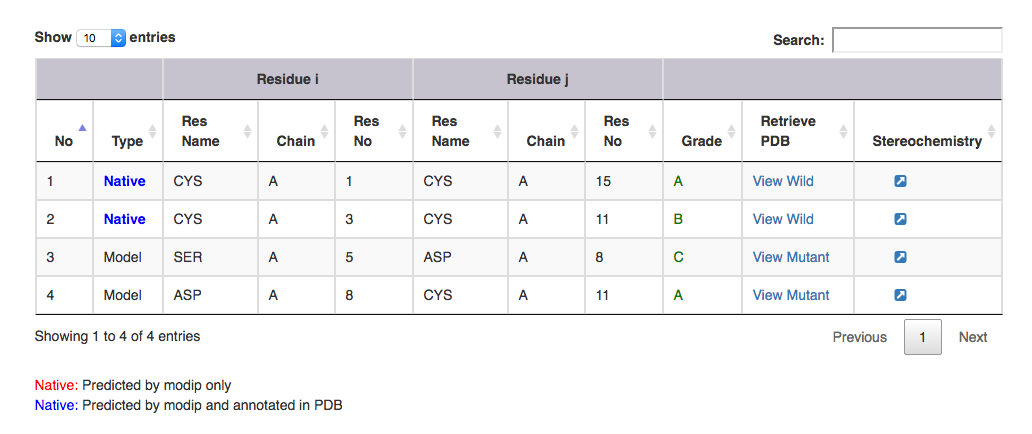
| Output features | Description |
|---|---|
| Type | gives information on whether it is a modelled or a native disulphide bridge. Annotated and unannotated SS bonds: where both the residues are cysteines. If marked in the PDB file, blue colour. If predicted by MODIP then native is represented in red colour. |
| Residue i | gives information on the start residue name, chain ID and residue number of the SS bond. |
| Residue j | gives information on the end residue name, chain ID and residue number of the SS bond. |
| Grade | grade distribution of modelled and native disulphides. |
| Retrive PDB | gives PDB file of the substructural segment with residue i and j mutated to cysteines ( Example: for Modelled SS bond, mutant PDB file is available with residue no 5 to 8 mutated to Cysteines ). User can view the segment in Jsmol and PDB file is available for download. |
| Stereochemistry | hyperlink to view the stereochemical parameters of SS bond. |
Downloadable Results
- Results: summary information. (txt file)
- Modelled Disulphide: Native and Modelled disulphide details (Txt file)
- Log File : Execution log
Details of MODIP procedure
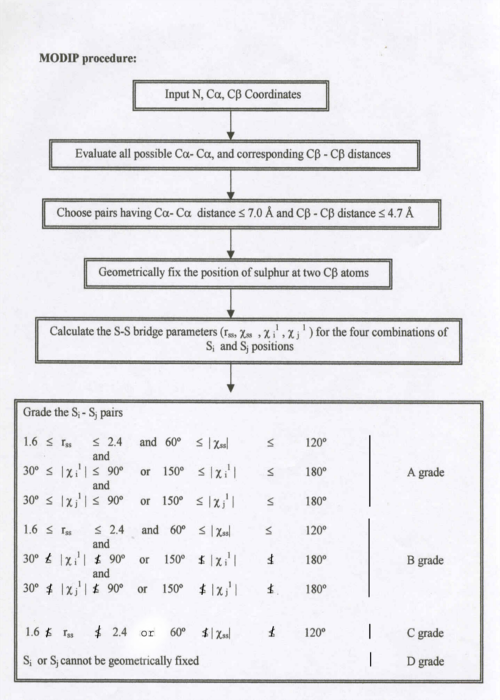
This computer modelling program requires N,C alpha ,C beta coordinates as input and considers all possible residue pairs and calculate the C(alpha)-C(alpha), C(beta)-C(beta) distances. It selects the residue pair with C(alpha)-C(alpha) distance of less than or equal to 6.5 Angstrom and C(beta)-C(beta) distance of less than or equal to 4.5 Angstrom and geometricaly fixes the sulphur atom and grades them based on the stereochemical quality.
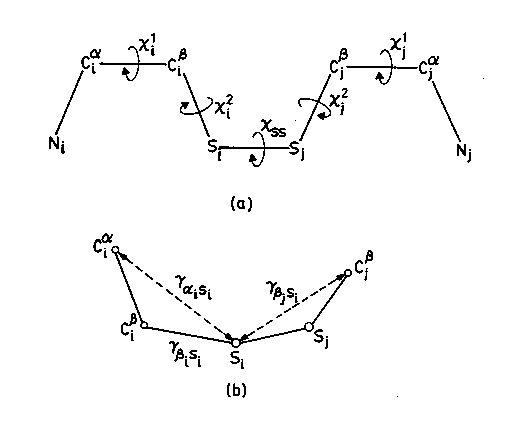
Details of various Dihedral angles(a) and inter atomic distances(b) for fixing Sulphur atoms
Summary of MODIP procedure
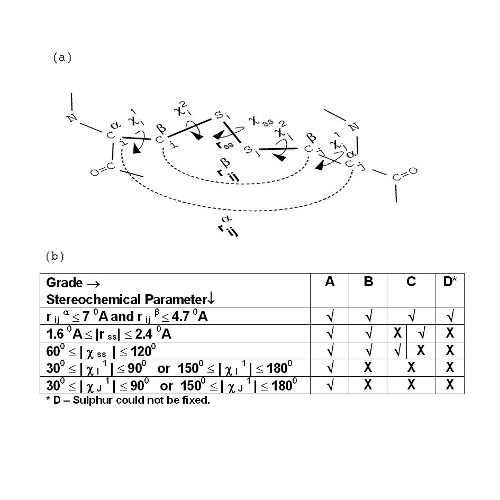
Explanation of grades
References
R.Sowdhamini, N.Srinivasan, B.Shoichet, D.V.Santi, C.Ramakrishnan and P.Balaram (1989). Stereochemical modelling of disulfide bridges: Criteria for introduction into proteins by site-directed mutagenesis. Prot. Engng., 3, 95-103.
R.R.Thangudu, A.Vinayagam, G.Pugalenthi, A.Manonmani, B.Offmann and R.Sowdhamini (2005). Native and modelled disulphide bonds in proteins: Knowledge-based approaches towards structure prediction of disulphide-rich polypeptides. Proteins: Struct., Funct., Bioinf., 58, 866-79.