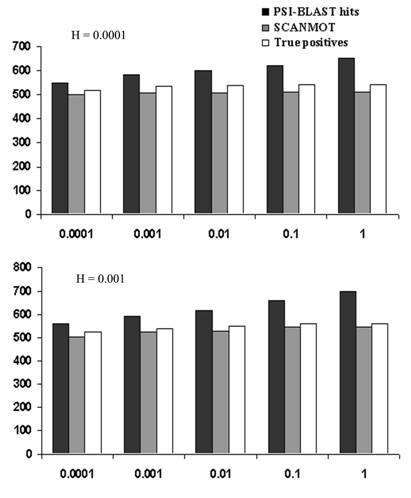
Validation of PSI-BLAST results:
SCANMOT
algorithm is employed to validate and characterize PSI-BLAST output to extract
true homologues. Following figure provides the number of homologous proteins identified
by PSI-BLAST runs considering each member of the nine families (list) as query sequence. Homologous
sequences are identified from a sequence database that contains closely related
sequences augmented for each SCOP (Murzin et al., 1995) protein entry whose
structure is known; this augmented sequence database provides information on
expected number of true positives. SCANMOT algorithm is applied on every
PSI-BLAST output and homologous sequences are identified on the basis of
presence of motifs, characteristic of the query family of proteins. The numbers
of proteins containing at least 50% of the total number of identified motifs
are regarded as significant homologues. The number of significant homologues
identified by SCANMOT motif scanning procedure is very much close to the actual
number of true positives. The observation of increasing number of false
positives with higher E-values supports earlier studies (Muller et al.,
1999). It has also been shown that the inclusion or H-value does not have much
effect on the number of true positives, as the numbers remain very similar for
different H-values. With increasing E-values, the number of hits obtained by
PSI-BLAST increases by 20 -25% whereas the increase in percentage of true
positives remains quite low (5 - 7%). The numbers and the inclusion rate (3 -
8%) of the significant homologues characterized by the presence of motifs are
also very similar to the true positives