|
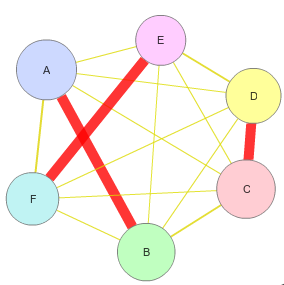
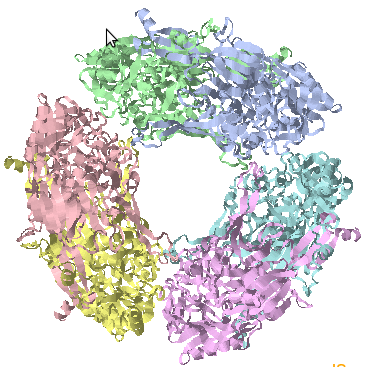
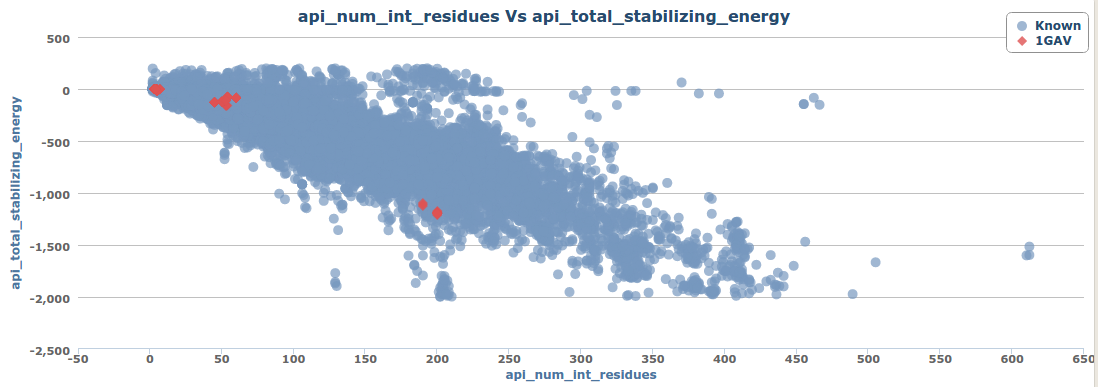
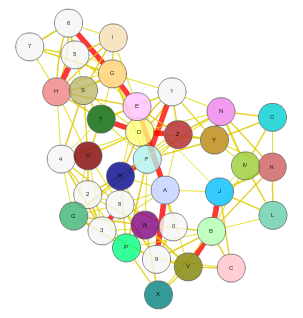
Cippa - Server for Chain Interactions in Protein-Protein Assemblies is a tool for analysing the inter chain interactions in a protein complex. This server considers all the possible combinations of chains in the structure file (.pdb) uploaded and identifies the true interactions based on various features such as Total stabilizing energy, Hydrogen Bonds, Electrostatic Energy, Interface Residues, Short Contacts, van der waals pairs and its energy, salt bridges etc. The results include the values calculated for each features considered and the graphical representation of the interactions and corresponding structure visualization. Cippa Server also provides CippaMap in which the energy values and interface residues from the query structure are mapped on to the values from the known complexes available in Proetein Data Bank (PDB). Cippa Server results are validation measures for your protein complexes modelled by any tool. Plotting the interaction from the complex on to the CippaMap provides more clarity on the correctness of the prediction. |
|
|
|
|
